当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
E2EDNA: Simulation Protocol for DNA Aptamers with Ligands
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-08-26 , DOI: 10.1021/acs.jcim.1c00696 Michael Kilgour 1 , Tao Liu 1 , Brandon D Walker 2 , Pengyu Ren 2 , Lena Simine 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-08-26 , DOI: 10.1021/acs.jcim.1c00696 Michael Kilgour 1 , Tao Liu 1 , Brandon D Walker 2 , Pengyu Ren 2 , Lena Simine 1
Affiliation
|
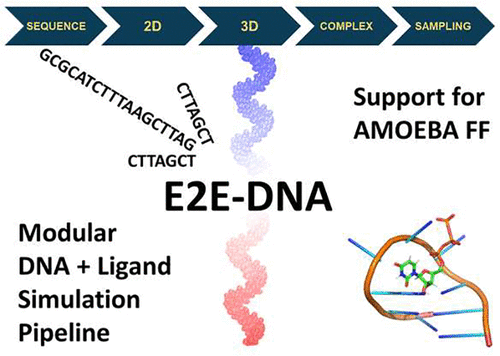
We present E2EDNA, a simulation protocol and accompanying code for the molecular biophysics and materials science communities. This protocol is both easy to use and sufficiently efficient to simulate single-stranded (ss)DNA and small analyte systems that are important to cellular processes and nanotechnologies such as DNA aptamer-based sensors. Existing computational tools used for aptamer design focus on cost-effective secondary structure prediction and motif analysis in the large data sets produced by SELEX experiments. As a rule, they do not offer flexibility with respect to the choice of the theoretical engine or direct access to the simulation platform. Practical aptamer optimization often requires higher accuracy predictions for only a small subset of sequences suggested, e.g., by SELEX experiments, but in the absence of a streamlined procedure, this task is extremely time and expertise intensive. We address this gap by introducing E2EDNA, a computational framework that accepts a DNA sequence in the FASTA format and the structures of the desired ligands and performs approximate folding followed by a refining step, analyte complexation, and molecular dynamics sampling at the desired level of accuracy. As a case study, we simulate a DNA-UTP (uridine triphosphate) complex in water using the state-of-the-art AMOEBA polarizable force field. The code is available at https://github.com/InfluenceFunctional/E2EDNA.
中文翻译:

E2EDNA:带有配体的 DNA 适配体的模拟协议
我们推出 E2EDNA,这是一种针对分子生物物理学和材料科学界的模拟协议和随附代码。该协议既易于使用又足够高效,可以模拟单链 (ss)DNA 和小型分析物系统,这些系统对于细胞过程和纳米技术(例如基于 DNA 适体的传感器)非常重要。用于适体设计的现有计算工具侧重于 SELEX 实验产生的大数据集中具有成本效益的二级结构预测和基序分析。通常,它们在选择理论引擎或直接访问仿真平台方面不提供灵活性。实际适体优化通常需要仅对一小部分序列进行更准确的预测,例如通过SELEX实验,但在缺乏简化程序的情况下,这项任务非常耗时和专业知识密集。我们通过引入 E2EDNA 来解决这一差距,E2EDNA 是一种计算框架,它接受 FASTA 格式的 DNA 序列和所需配体的结构,并执行近似折叠,然后以所需的精度水平进行精炼步骤、分析物络合和分子动力学采样。作为案例研究,我们使用最先进的 AMOEBA 极化力场模拟水中的 DNA-UTP(尿苷三磷酸)复合物。该代码可从 https://github.com/InfluenceFunctional/E2EDNA 获取。
更新日期:2021-09-27
中文翻译:

E2EDNA:带有配体的 DNA 适配体的模拟协议
我们推出 E2EDNA,这是一种针对分子生物物理学和材料科学界的模拟协议和随附代码。该协议既易于使用又足够高效,可以模拟单链 (ss)DNA 和小型分析物系统,这些系统对于细胞过程和纳米技术(例如基于 DNA 适体的传感器)非常重要。用于适体设计的现有计算工具侧重于 SELEX 实验产生的大数据集中具有成本效益的二级结构预测和基序分析。通常,它们在选择理论引擎或直接访问仿真平台方面不提供灵活性。实际适体优化通常需要仅对一小部分序列进行更准确的预测,例如通过SELEX实验,但在缺乏简化程序的情况下,这项任务非常耗时和专业知识密集。我们通过引入 E2EDNA 来解决这一差距,E2EDNA 是一种计算框架,它接受 FASTA 格式的 DNA 序列和所需配体的结构,并执行近似折叠,然后以所需的精度水平进行精炼步骤、分析物络合和分子动力学采样。作为案例研究,我们使用最先进的 AMOEBA 极化力场模拟水中的 DNA-UTP(尿苷三磷酸)复合物。该代码可从 https://github.com/InfluenceFunctional/E2EDNA 获取。











































 京公网安备 11010802027423号
京公网安备 11010802027423号