Molecular Cell ( IF 14.5 ) Pub Date : 2021-08-26 , DOI: 10.1016/j.molcel.2021.07.043 Renjian Xiao 1 , Shukun Wang 1 , Ruijie Han 1 , Zhuang Li 1 , Clinton Gabel 1 , Indranil Arun Mukherjee 1 , Leifu Chang 2
|
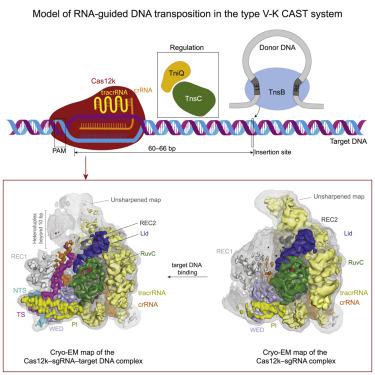
The type V-K CRISPR-Cas system, featured by Cas12k effector with a naturally inactivated RuvC domain and associated with Tn7-like transposon for RNA-guided DNA transposition, is a promising tool for precise DNA insertion. To reveal the mechanism underlying target DNA recognition, we determined a cryoelectron microscopy (cryo-EM) structure of Cas12k from cyanobacteria Scytonema hofmanni in complex with a single guide RNA (sgRNA) and a double-stranded target DNA. Coupled with mutagenesis and in vitro DNA transposition assay, our results revealed mechanisms for the recognition of the GGTT protospacer adjacent motif (PAM) sequence and the structural elements of Cas12k critical for RNA-guided DNA transposition. These structural and mechanistic insights should aid in the development of type V-K CRISPR-transposon systems as tools for genome editing.
中文翻译:

CRISPR-Cas12k 识别靶标 DNA 进行 RNA 引导 DNA 转座的结构基础
VK 型 CRISPR-Cas 系统的特点是具有天然失活的 RuvC 结构域的 Cas12k 效应子,并与用于 RNA 引导 DNA 转座的 Tn7 样转座子相关,是一种有前途的精确 DNA 插入工具。为了揭示目标 DNA 识别的机制,我们确定了来自蓝藻Scytonema hofmanni的 Cas12k 与单引导 RNA (sgRNA) 和双链目标 DNA 复合物的冷冻电子显微镜 (cryo-EM) 结构。结合诱变和体外DNA 转座测定,我们的结果揭示了 GGTT 原型间隔子相邻基序 (PAM) 序列的识别机制以及对 RNA 引导的 DNA 转座至关重要的 Cas12k 结构元件。这些结构和机制见解应有助于开发 VK 型 CRISPR 转座子系统作为基因组编辑工具。











































 京公网安备 11010802027423号
京公网安备 11010802027423号