当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Expression of Alternative Nitrogenases in Rhodopseudomonas palustris Is Enhanced Using an Optimized Genetic Toolset for Rapid, Markerless Modifications
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-08-25 , DOI: 10.1021/acssynbio.0c00496 Jan-Pierre du Toit 1 , David J Lea-Smith 2, 3 , Anna Git 3 , John R D Hervey 3 , Christopher J Howe 3 , Robert W M Pott 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-08-25 , DOI: 10.1021/acssynbio.0c00496 Jan-Pierre du Toit 1 , David J Lea-Smith 2, 3 , Anna Git 3 , John R D Hervey 3 , Christopher J Howe 3 , Robert W M Pott 1
Affiliation
|
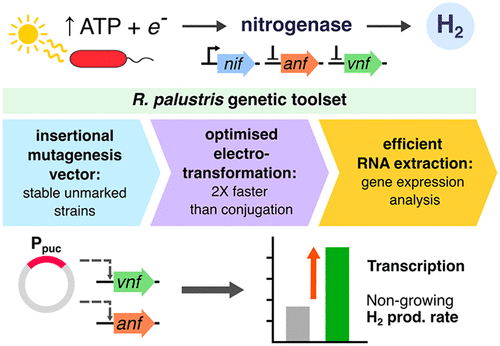
The phototrophic bacterium Rhodopseudomonas palustris is emerging as a promising biotechnological chassis organism, due to its resilience to a range of harsh conditions, a wide metabolic repertoire, and the ability to quickly regenerate ATP using light. However, realization of this promise is impeded by a lack of efficient, rapid methods for genetic modification. Here, we present optimized tools for generating chromosomal insertions and deletions employing electroporation as a means of transformation. Generation of markerless strains can be completed in 12 days, approximately half the time for previous conjugation-based methods. This system was used for overexpression of alternative nitrogenase isozymes with the aim of improving biohydrogen productivity. Insertion of the pucBa promoter upstream of vnf and anf nitrogenase operons drove robust overexpression up to 4000-fold higher than wild-type. Transcript quantification was facilitated by an optimized high-quality RNA extraction protocol employing lysis using detergent and heat. Overexpression resulted in increased nitrogenase protein levels, extending to superior hydrogen productivity in bioreactor studies under nongrowing conditions, where promoter-modified strains better utilized the favorable energy state created by reduced competition from cell division. Robust heterologous expression driven by the pucBa promoter is thus attractive for energy-intensive biosyntheses suited to the capabilities of R. palustris. Development of this genetic modification toolset will accelerate the advancement of R. palustris as a biotechnological chassis organism, and insights into the effects of nitrogenase overexpression will guide future efforts in engineering strains for improved hydrogen production.
中文翻译:

使用优化的遗传工具集进行快速、无标记修饰,增强了沼泽红假单胞菌中替代固氮酶的表达
光养细菌Rhodopseudomonas palustris正在成为一种有前途的生物技术底盘生物,因为它具有对一系列恶劣条件的适应能力、广泛的代谢库以及利用光快速再生 ATP 的能力。然而,由于缺乏有效、快速的基因改造方法,这一承诺的实现受到了阻碍。在这里,我们提出了使用电穿孔作为转化手段来生成染色体插入和缺失的优化工具。无标记菌株的生成可在 12 天内完成,大约是以前基于缀合方法的一半时间。该系统用于替代固氮酶同工酶的过表达,目的是提高生物氢的生产力。pucBa的插入vnf和anf固氮酶操纵子上游的启动子驱动强过表达比野生型高 4000 倍。优化的高质量 RNA 提取方案促进了转录物的量化,该方案采用使用洗涤剂和加热的裂解。过表达导致固氮酶蛋白水平增加,在非生长条件下的生物反应器研究中延伸到更高的氢生产力,其中启动子修饰的菌株更好地利用了细胞分裂竞争减少所产生的有利能量状态。因此,由pucBa启动子驱动的稳健异源表达对于适合R. palustris能力的能量密集型生物合成具有吸引力. 这种基因改造工具集的开发将加速R. palustris作为生物技术底盘生物的发展,对固氮酶过度表达影响的深入了解将指导未来在工程菌株方面的努力,以提高产氢能力。
更新日期:2021-09-17
中文翻译:

使用优化的遗传工具集进行快速、无标记修饰,增强了沼泽红假单胞菌中替代固氮酶的表达
光养细菌Rhodopseudomonas palustris正在成为一种有前途的生物技术底盘生物,因为它具有对一系列恶劣条件的适应能力、广泛的代谢库以及利用光快速再生 ATP 的能力。然而,由于缺乏有效、快速的基因改造方法,这一承诺的实现受到了阻碍。在这里,我们提出了使用电穿孔作为转化手段来生成染色体插入和缺失的优化工具。无标记菌株的生成可在 12 天内完成,大约是以前基于缀合方法的一半时间。该系统用于替代固氮酶同工酶的过表达,目的是提高生物氢的生产力。pucBa的插入vnf和anf固氮酶操纵子上游的启动子驱动强过表达比野生型高 4000 倍。优化的高质量 RNA 提取方案促进了转录物的量化,该方案采用使用洗涤剂和加热的裂解。过表达导致固氮酶蛋白水平增加,在非生长条件下的生物反应器研究中延伸到更高的氢生产力,其中启动子修饰的菌株更好地利用了细胞分裂竞争减少所产生的有利能量状态。因此,由pucBa启动子驱动的稳健异源表达对于适合R. palustris能力的能量密集型生物合成具有吸引力. 这种基因改造工具集的开发将加速R. palustris作为生物技术底盘生物的发展,对固氮酶过度表达影响的深入了解将指导未来在工程菌株方面的努力,以提高产氢能力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号