当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Solving 0–1 Integer Programming Problem Based on DNA Strand Displacement Reaction Network
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-08-25 , DOI: 10.1021/acssynbio.1c00244 Zhen Tang 1 , Zhixiang Yin 1, 2 , Luhui Wang 3 , Jianzhong Cui 4 , Jing Yang 1 , Risheng Wang 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-08-25 , DOI: 10.1021/acssynbio.1c00244 Zhen Tang 1 , Zhixiang Yin 1, 2 , Luhui Wang 3 , Jianzhong Cui 4 , Jing Yang 1 , Risheng Wang 1
Affiliation
|
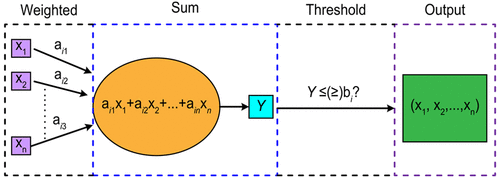
Chemical reaction networks (CRNs) based on DNA strand displacement (DSD) can be used as an effective programming language for solving various mathematical problems. In this paper, we design three chemical reaction modules by using the DNA strand displacement reaction as the basic principle, with a weighted reaction module, sum reaction module, and threshold reaction module. These modules are used as basic elements to form chemical reaction networks that can be used to solve 0–1 integer programming problems. The problem can be solved through the three steps of weighting, sum, and threshold, and then the results of the operations can be expressed through a single-stranded DNA output with fluorescent molecules. Finally, we use biochemical experiments and Visual DSD simulation software to verify and evaluate the chemical reaction networks. The results have shown that the DSD-based chemical reaction networks constructed in this paper have good feasibility and stability.
中文翻译:

基于DNA链置换反应网络求解0-1整数规划问题
基于 DNA 链置换 (DSD) 的化学反应网络 (CRN) 可用作解决各种数学问题的有效编程语言。本文以DNA链置换反应为基本原理,设计了三个化学反应模块,分别为加权反应模块、总和反应模块和阈值反应模块。这些模块用作基本元素来形成可用于解决 0-1 整数规划问题的化学反应网络。这个问题可以通过加权、求和、阈值三个步骤来解决,然后运算的结果可以通过带有荧光分子的单链DNA输出来表达。最后,我们使用生化实验和Visual DSD模拟软件来验证和评估化学反应网络。
更新日期:2021-09-17
中文翻译:

基于DNA链置换反应网络求解0-1整数规划问题
基于 DNA 链置换 (DSD) 的化学反应网络 (CRN) 可用作解决各种数学问题的有效编程语言。本文以DNA链置换反应为基本原理,设计了三个化学反应模块,分别为加权反应模块、总和反应模块和阈值反应模块。这些模块用作基本元素来形成可用于解决 0-1 整数规划问题的化学反应网络。这个问题可以通过加权、求和、阈值三个步骤来解决,然后运算的结果可以通过带有荧光分子的单链DNA输出来表达。最后,我们使用生化实验和Visual DSD模拟软件来验证和评估化学反应网络。











































 京公网安备 11010802027423号
京公网安备 11010802027423号