当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-08-21 , DOI: 10.1016/j.csbj.2021.08.030 Anders Nørgaard Sørensen 1 , Cedric Woudstra 1 , Martine C Holst Sørensen 1 , Lone Brøndsted 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-08-21 , DOI: 10.1016/j.csbj.2021.08.030 Anders Nørgaard Sørensen 1 , Cedric Woudstra 1 , Martine C Holst Sørensen 1 , Lone Brøndsted 1
Affiliation
|
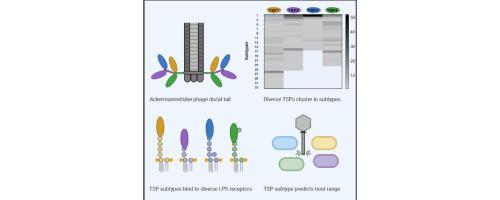
Phages belonging to the family encode up to four tail spike proteins (TSPs), each recognizing a specific receptor of their bacterial hosts. Here, we determined the TSPs diversity of 99 phages by performing a comprehensive analysis. Based on sequence diversity, we assigned all TSPs into distinctive subtypes of TSP1, TSP2, TSP3 and TSP4, and found each TSP subtype to be specifically associated with the genera (, , ) of the family. Further analysis showed that the N-terminal XD1 and XD2 domains in TSP2 and TSP4, hinging the four TSPs together, are preserved. In contrast, the C-terminal receptor binding modules were only conserved within TSP subtypes, except for some TSP1s and TSP3s that were similar to specific TSP4s. A conserved motif in TSP1, TSP3 and TSP4 of phages may allow recombination between receptor binding modules, thus altering host recognition. The receptors for numerous uncharacterized phages expressing TSPs in the same subtypes were predicted using previous host range data. To validate our predictions, we experimentally determined the host recognition of three of the four TSPs expressed by kuttervirus S117. We confirmed that S117 TSP1 and TSP2 bind to their predicted host receptors, and identified the receptor for TSP3, which is shared by 51 other phages. phages were thus shown encode a vast genetic diversity of potentially exchangeable TSPs influencing host recognition. Overall, our study demonstrates that comprehensive and host range analysis of TSPs can predict host recognition of phages.
中文翻译:

尾部刺突蛋白的亚型预测阿克曼病毒科噬菌体的宿主范围
该家族的噬菌体编码多达四种尾部刺突蛋白(TSP),每种蛋白都能识别其细菌宿主的特定受体。在这里,我们通过综合分析确定了 99 个噬菌体的 TSP 多样性。基于序列多样性,我们将所有 TSP 分为不同的亚型 TSP1、TSP2、TSP3 和 TSP4,并发现每个 TSP 亚型与该科的属 (, , ) 特异性相关。进一步分析表明,TSP2 和 TSP4 中将四个 TSP 连接在一起的 N 端 XD1 和 XD2 结构域得以保留。相比之下,除了一些与特定 TSP4 相似的 TSP1 和 TSP3 之外,C 端受体结合模块仅在 TSP 亚型中保守。噬菌体 TSP1、TSP3 和 TSP4 中的保守基序可能允许受体结合模块之间发生重组,从而改变宿主识别。使用先前的宿主范围数据预测了许多表达相同亚型 TSP 的未表征噬菌体的受体。为了验证我们的预测,我们通过实验确定了 kuttervirus S117 表达的四个 TSP 中的三个的宿主识别。我们确认 S117 TSP1 和 TSP2 与其预测的宿主受体结合,并鉴定了 TSP3 的受体,该受体为 51 个其他噬菌体所共有。因此,噬菌体编码了影响宿主识别的潜在可交换 TSP 的巨大遗传多样性。总的来说,我们的研究表明,对 TSP 进行全面的宿主范围分析可以预测噬菌体的宿主识别。
更新日期:2021-08-21
中文翻译:

尾部刺突蛋白的亚型预测阿克曼病毒科噬菌体的宿主范围
该家族的噬菌体编码多达四种尾部刺突蛋白(TSP),每种蛋白都能识别其细菌宿主的特定受体。在这里,我们通过综合分析确定了 99 个噬菌体的 TSP 多样性。基于序列多样性,我们将所有 TSP 分为不同的亚型 TSP1、TSP2、TSP3 和 TSP4,并发现每个 TSP 亚型与该科的属 (, , ) 特异性相关。进一步分析表明,TSP2 和 TSP4 中将四个 TSP 连接在一起的 N 端 XD1 和 XD2 结构域得以保留。相比之下,除了一些与特定 TSP4 相似的 TSP1 和 TSP3 之外,C 端受体结合模块仅在 TSP 亚型中保守。噬菌体 TSP1、TSP3 和 TSP4 中的保守基序可能允许受体结合模块之间发生重组,从而改变宿主识别。使用先前的宿主范围数据预测了许多表达相同亚型 TSP 的未表征噬菌体的受体。为了验证我们的预测,我们通过实验确定了 kuttervirus S117 表达的四个 TSP 中的三个的宿主识别。我们确认 S117 TSP1 和 TSP2 与其预测的宿主受体结合,并鉴定了 TSP3 的受体,该受体为 51 个其他噬菌体所共有。因此,噬菌体编码了影响宿主识别的潜在可交换 TSP 的巨大遗传多样性。总的来说,我们的研究表明,对 TSP 进行全面的宿主范围分析可以预测噬菌体的宿主识别。









































 京公网安备 11010802027423号
京公网安备 11010802027423号