Computational and Structural Biotechnology Journal ( IF 6 ) Pub Date : 2021-08-19 , DOI: 10.1016/j.csbj.2021.08.023 Ma'mon M Hatmal 1 , Omar Abuyaman 1 , Mutasem Taha 2
|
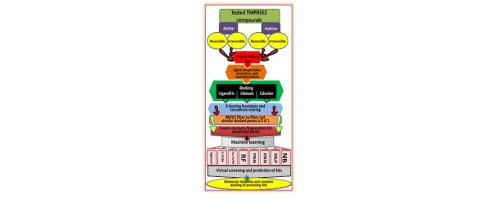
In the present work we introduce the use of multiple docked poses for bootstrapping machine learning-based QSAR modelling. Ligand-receptor contact fingerprints are implemented as descriptor variables. We implemented this method for the discovery of potential inhibitors of the serine protease enzyme TMPRSS2 involved the infectivity of coronaviruses. Several machine learners were scanned, however, Xgboost, support vector machines (SVM) and random forests (RF) were the best with testing set accuracies reaching 90%. Three potential hits were identified upon using the method to scan known untested FDA approved drugs against TMPRSS2. Subsequent molecular dynamics simulation and covalent docking supported the results of the new computational approach.
中文翻译:

用于引导生物活性分类机器学习的对接生成的多个配体姿势:重新利用共价抑制剂用于与 COVID-19 相关的 TMPRSS2 作为案例研究
在目前的工作中,我们介绍了使用多个对接姿势来引导基于机器学习的 QSAR 建模。配体-受体接触指纹被实现为描述符变量。我们采用这种方法来发现与冠状病毒感染性相关的丝氨酸蛋白酶 TMPRSS2 的潜在抑制剂。扫描了多个机器学习器,但 Xgboost、支持向量机 (SVM) 和随机森林 (RF) 是最好的,测试集准确率达到 90%。使用该方法扫描已知的未经 FDA 批准的针对 TMPRSS2 的药物后,发现了三个潜在的命中。随后的分子动力学模拟和共价对接支持了新计算方法的结果。



























 京公网安备 11010802027423号
京公网安备 11010802027423号