当前位置:
X-MOL 学术
›
J. Phys. Chem. B
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Unraveling the Allosteric Mechanism of Four Cancer-related Mutations in the Disruption of p53-DNA Interaction
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2021-08-17 , DOI: 10.1021/acs.jpcb.1c05638 Yiming Tang 1 , Yifei Yao 1 , Guanghong Wei 1
The Journal of Physical Chemistry B ( IF 2.8 ) Pub Date : 2021-08-17 , DOI: 10.1021/acs.jpcb.1c05638 Yiming Tang 1 , Yifei Yao 1 , Guanghong Wei 1
Affiliation
|
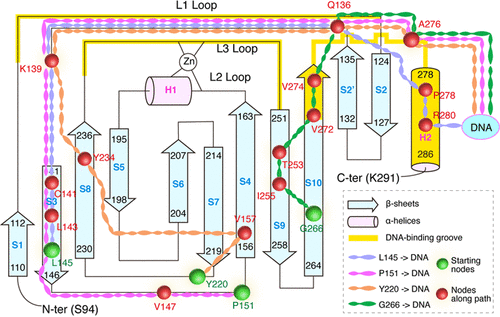
The p53 protein plays active roles in the physiological regulation of cell cycle as well as in cancer developments. In more than half of human cancers, the protein is inactivated by mutations located primarily in its DNA-binding domain (DBD), and some mutations located in the β-sandwich region of DBD are reported to decrease p53–DNA binding affinities. To understand the long-range correlation between p53 β-sandwich and DNA, and the allosteric mechanism of β-sandwich mutations in the disruption of p53–DNA interactions, we first identify three regions with a strong correlation with DNA based on microsecond molecular dynamics (MD) simulations of wild-type p53–DNA complex and then perform multiple MD simulations on four cancer-related mutants L145Q, P151S, Y220C, and G266R, which are located in these three regions. Our simulations show that these mutations allosterically destabilize the structural stability of the DNA-binding groove in p53 and disrupt the p53–DNA interactions. Network analyses reveal optimal correlation paths through which the mutation-induced allosteric signal passes to DNA, and the disturbance effect of these mutations on the global connectivity and dynamical correlation of the p53–DNA complex. This work paves the way for the in-depth understanding of the mutation-induced loss in p53’s DNA-recognition ability and the pathological mechanism of cancer development.
中文翻译:

揭示四种癌症相关突变破坏 p53-DNA 相互作用的变构机制
p53 蛋白在细胞周期的生理调节以及癌症发展中发挥着积极作用。在超过一半的人类癌症中,该蛋白质因主要位于其 DNA 结合域 (DBD) 的突变而失活,据报道,位于 DBD 的 β-夹心区的一些突变会降低 p53-DNA 结合亲和力。为了了解 p53 β-sandwich 与 DNA 之间的长程相关性,以及 β-sandwich 突变在破坏 p53-DNA 相互作用中的变构机制,我们首先基于微秒分子动力学确定了与 DNA 具有强相关性的三个区域。 MD) 模拟野生型 p53-DNA 复合物,然后对位于这三个区域的四种癌症相关突变体 L145Q、P151S、Y220C 和 G266R 进行多次 MD 模拟。我们的模拟表明,这些突变变构破坏了 p53 中 DNA 结合槽的结构稳定性并破坏了 p53-DNA 相互作用。网络分析揭示了突变诱导的变构信号传递到 DNA 的最佳相关路径,以及这些突变对 p53-DNA 复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。以及这些突变对p53-DNA复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。以及这些突变对p53-DNA复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。
更新日期:2021-09-16
中文翻译:

揭示四种癌症相关突变破坏 p53-DNA 相互作用的变构机制
p53 蛋白在细胞周期的生理调节以及癌症发展中发挥着积极作用。在超过一半的人类癌症中,该蛋白质因主要位于其 DNA 结合域 (DBD) 的突变而失活,据报道,位于 DBD 的 β-夹心区的一些突变会降低 p53-DNA 结合亲和力。为了了解 p53 β-sandwich 与 DNA 之间的长程相关性,以及 β-sandwich 突变在破坏 p53-DNA 相互作用中的变构机制,我们首先基于微秒分子动力学确定了与 DNA 具有强相关性的三个区域。 MD) 模拟野生型 p53-DNA 复合物,然后对位于这三个区域的四种癌症相关突变体 L145Q、P151S、Y220C 和 G266R 进行多次 MD 模拟。我们的模拟表明,这些突变变构破坏了 p53 中 DNA 结合槽的结构稳定性并破坏了 p53-DNA 相互作用。网络分析揭示了突变诱导的变构信号传递到 DNA 的最佳相关路径,以及这些突变对 p53-DNA 复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。以及这些突变对p53-DNA复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。以及这些突变对p53-DNA复合物的全局连通性和动态相关性的干扰作用。这项工作为深入了解突变引起的p53 DNA识别能力丧失和癌症发展的病理机制铺平了道路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号