Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2021-08-17 , DOI: 10.1016/j.jmgm.2021.108008 Anna Antoniak 1 , Iga Biskupek 1 , Krzysztof K Bojarski 1 , Cezary Czaplewski 1 , Artur Giełdoń 1 , Mateusz Kogut 1 , Małgorzata M Kogut 1 , Paweł Krupa 2 , Agnieszka G Lipska 1 , Adam Liwo 3 , Emilia A Lubecka 4 , Mateusz Marcisz 5 , Martyna Maszota-Zieleniak 1 , Sergey A Samsonov 1 , Adam K Sieradzan 1 , Magdalena J Ślusarz 1 , Rafał Ślusarz 1 , Patryk A Wesołowski 5 , Karolina Ziȩba 1
|
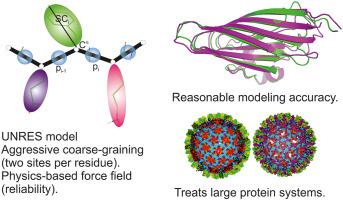
The UNited RESidue (UNRES) force field was tested in the 14th Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction (CASP14), in which larger oligomeric and multimeric targets were present compared to previous editions. Three prediction modes were tested (i) ab initio (the UNRES group), (ii) contact-assisted (the UNRES-contact group), and (iii) template-assisted (the UNRES-template group). For most of the targets, the contact restraints were derived from the server models top-ranked by the DeepQA method, while the DNCON2 method was used for 11 targets. Our consensus-fragment procedure was used to run template-assisted predictions. Each group also processed the Nuclear Magnetic Resonance (NMR)- and Small Angle X-Ray Scattering (SAXS)-data assisted targets. The average Global Distance Test Total Score (GDT_TS) of the ‘Model 1’ predictions were 29.17, 39.32, and 56.37 for the UNRES, UNRES-contact, and UNRES-template predictions, respectively, increasing by 0.53, 2.24, and 3.76, respectively, compared to CASP13. It was also found that the GDT_TS of the UNRES models obtained in ab initio mode and in the contact-assisted mode decreases with the square root of chain length, while the exponent in this relationship is 0.20 for the UNRES-template group models and 0.11 for the best performing AlphaFold2 models, which suggests that incorporation of database information, which stems from protein evolution, brings in long-range correlations, thus enabling the correction of force-field inaccuracies.
中文翻译:

在 CASP14 实验中使用粗粒度 UNRES 力场对蛋白质结构进行建模
UNITED RESidue (UNRES) 力场在第 14 次社区范围的蛋白质结构预测技术关键评估实验 (CASP14) 中进行了测试,其中与以前的版本相比,存在更大的寡聚体和多聚体目标。测试了三种预测模式 (i)从头开始(UNRES 组),(ii) 接触辅助(UNRES 接触组),和 (iii) 模板辅助(UNRES 模板组)。对于大多数目标,接触约束来自 DeepQA 方法排名靠前的服务器模型,而 DNCON2 方法用于 11 个目标。我们的共识片段程序用于运行模板辅助预测。每组还处理了核磁共振 (NMR) 和小角度 X 射线散射 (SAXS) 数据辅助目标。UNRES、UNRES-contact 和 UNRES-template 预测的“模型 1”预测的平均 Global Distance Test Total Score (GDT_TS) 分别为 29.17、39.32 和 56.37,分别增加了 0.53、2.24 和 3.76 ,与 CASP13 相比。还发现从头计算得到的 UNRES 模型的 GDT_TS 模式和接触辅助模式随着链长的平方根而减小,而这种关系的指数对于 UNRES 模板组模型为 0.20,对于性能最好的 AlphaFold2 模型为 0.11,这表明数据库信息的合并,源于蛋白质进化,带来长期相关性,从而能够纠正力场的不准确性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号