Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2021-08-17 , DOI: 10.1016/j.jmgm.2021.108007 Ana Virginia Frota Guimarães 1 , Natália Fernandes Frota 2 , Marcos Roberto Lourenzoni 2
|
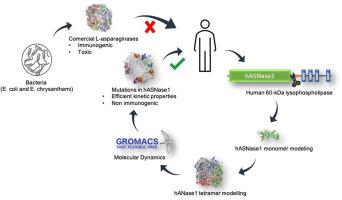
The l-asparaginase enzyme is used in cancer therapy, mainly acute lymphoid leukemia (ALL). Commercial enzymes (EcASNase2) cause adverse reactions during treatment, such as immunogenicity. A human enzyme could be a non-immunogenic substitute. However, no candidate was found showing efficient kinetic properties. HASNase1 is an l-asparaginase that comes from the N-terminal domain of a protein called 60 kDa-lysophospholipase and its 3D structure has not been resolved. HASNase1 is homologous to EcASNase1 and gpASNase1, and this last one has shown efficient kinetic properties. Homology modeling was used to find the 3D structure of hASNase1, so one could submit it to Molecular Dynamics (MD), in order to understand structural differences that lead to different catalytic efficiency compared to EcASNase2 and gpASNase1. The interaction potential between L-Asn and active site residues showed that the substrate can rotate in the site when Region1 is open. Region1 residues sequence favors deformations and movements as shown in MD. Region2-A is linear in gpASNase1, and it features a helix portion in hASNase1, which leaves the Tyr308 position projected to the active site ratifying its role in catalytic efficiency. Analysis of Lys188 orientation and movement showed the effect of positive cooperativity in hASNase1. It was found that the presence of Asn at the allosteric site helps, not only in Region1 stabilization, but also in Lys188 stabilization for the maintenance of the triad. Despite structural similarities in hASNase1, gpASNase1, and EcASNase2, there are differences in structural determinants that, in addition to allosterism, may explain the different kinetic properties.
中文翻译:

人 L-天冬酰胺酶 1 的分子动力学模拟:对酶活性结构决定因素的洞察
L-天冬酰胺酶用于癌症治疗,主要是急性淋巴细胞白血病 (ALL)。商业酶(Ec ASNase2)在治疗过程中会引起不良反应,例如免疫原性。人类酶可能是一种非免疫原性替代品。然而,没有发现显示出有效动力学特性的候选物。HASNase1 是一种l-天冬酰胺酶,来自称为 60 kDa-溶血磷脂酶的蛋白质的 N 端结构域,其 3D 结构尚未解析。HASNase1 与Ec同源ASNase1 和 gpASNase1,最后一个显示出有效的动力学特性。同源建模用于查找 hASNase1 的 3D 结构,因此可以将其提交给分子动力学 (MD),以了解与Ec相比导致不同催化效率的结构差异ASNase2 和 gpASNase1。L-Asn 与活性位点残基之间的相互作用势表明,当 Region1 打开时,底物可以在位点内旋转。Region1 残基序列有利于变形和运动,如 MD 所示。Region2-A 在 gpASNase1 中是线性的,它在 hASNase1 中具有螺旋部分,这使得 Tyr308 位置投射到活性位点,从而证明了其在催化效率中的作用。Lys188 方向和运动的分析显示了在 hASNase1 中的正协同作用。发现变构位点 Asn 的存在不仅有助于区域 1 的稳定,而且有助于维持三元组的 Lys188 稳定。尽管 hASNase1、gpASNase1 和Ec的结构相似ASNase2,除了变构之外,结构决定因素存在差异,可以解释不同的动力学特性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号