当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
BiasNet: A Model to Predict Ligand Bias Toward GPCR Signaling
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-08-16 , DOI: 10.1021/acs.jcim.1c00317 Jason E Sanchez 1 , Govinda B Kc 1 , Julian Franco 2 , William J Allen 3 , Jesus David Garcia 4 , Suman Sirimulla 1, 4, 5
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-08-16 , DOI: 10.1021/acs.jcim.1c00317 Jason E Sanchez 1 , Govinda B Kc 1 , Julian Franco 2 , William J Allen 3 , Jesus David Garcia 4 , Suman Sirimulla 1, 4, 5
Affiliation
|
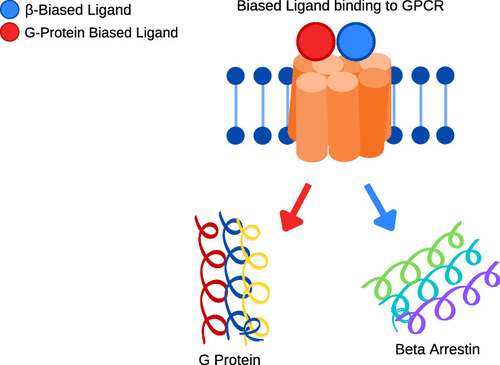
Signaling bias is a feature of many G protein-coupled receptor (GPCR) targeting drugs with potential clinical implications. Whether it is therapeutically advantageous for a drug to be G protein biased or β-arrestin biased depends on the context of the signaling pathway. Here, we explored GPCR ligands that exhibit biased signaling to gain insights into scaffolds and pharmacophores that lead to bias. More specifically, we considered BiasDB, a database containing information about GPCR biased ligands, and focused our analysis on ligands which show either a G protein or β-arrestin bias. Five different machine learning models were trained on these ligands using 15 different sets of features. Molecular fragments which were important for training the models were analyzed. Two of these fragments (number of secondary amines and number of aromatic amines) were more prevalent in β-arrestin biased ligands. After training a random forest model on HierS scaffolds, we found five scaffolds, which demonstrated G protein or β-arrestin bias. We also conducted t-SNE clustering, observing correspondence between unsupervised and supervised machine learning methods. To increase the applicability of our work, we developed a web implementation of our models, which can predict bias based on user-provided SMILES, drug names, or PubChem CID. Our web implementation is available at: drugdiscovery.utep.edu/biasnet.
中文翻译:

BiasNet:预测配体对 GPCR 信号的偏差的模型
信号偏倚是许多具有潜在临床意义的 G 蛋白偶联受体 (GPCR) 靶向药物的特征。偏向 G 蛋白或偏向 β-抑制蛋白的药物在治疗上是否有利取决于信号通路的背景。在这里,我们探索了表现出偏向信号传导的 GPCR 配体,以深入了解导致偏向的支架和药效团。更具体地说,我们考虑了 BiasDB,一个包含有关 GPCR 偏向配体信息的数据库,并将我们的分析重点放在显示 G 蛋白或 β-抑制蛋白偏倚的配体上。使用 15 组不同的特征在这些配体上训练了五种不同的机器学习模型。分析了对训练模型很重要的分子片段。这些片段中的两个(仲胺的数量和芳香胺的数量)在β-抑制蛋白偏向配体中更为普遍。在 HierS 支架上训练随机森林模型后,我们发现了 5 个支架,它们证明了 G 蛋白或 β-arrestin 偏差。我们还进行了 t-SNE 聚类,观察无监督和有监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。观察无监督和监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。观察无监督和监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。
更新日期:2021-09-27
中文翻译:

BiasNet:预测配体对 GPCR 信号的偏差的模型
信号偏倚是许多具有潜在临床意义的 G 蛋白偶联受体 (GPCR) 靶向药物的特征。偏向 G 蛋白或偏向 β-抑制蛋白的药物在治疗上是否有利取决于信号通路的背景。在这里,我们探索了表现出偏向信号传导的 GPCR 配体,以深入了解导致偏向的支架和药效团。更具体地说,我们考虑了 BiasDB,一个包含有关 GPCR 偏向配体信息的数据库,并将我们的分析重点放在显示 G 蛋白或 β-抑制蛋白偏倚的配体上。使用 15 组不同的特征在这些配体上训练了五种不同的机器学习模型。分析了对训练模型很重要的分子片段。这些片段中的两个(仲胺的数量和芳香胺的数量)在β-抑制蛋白偏向配体中更为普遍。在 HierS 支架上训练随机森林模型后,我们发现了 5 个支架,它们证明了 G 蛋白或 β-arrestin 偏差。我们还进行了 t-SNE 聚类,观察无监督和有监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。观察无监督和监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。观察无监督和监督机器学习方法之间的对应关系。为了提高我们工作的适用性,我们开发了我们模型的网络实现,它可以根据用户提供的 SMILES、药物名称或 PubChem CID 预测偏差。我们的网络实施可在以下网址获得:drugdiscovery.utep.edu/biasnet。











































 京公网安备 11010802027423号
京公网安备 11010802027423号