Biochimica et Biophysica Acta (BBA) - General Subjects ( IF 2.8 ) Pub Date : 2021-08-12 , DOI: 10.1016/j.bbagen.2021.129990 Xiangyu Zhang 1 , Yixiang Sun 2 , Ziheng Zhang 2 , Hanxun Wang 1 , Jian Wang 1 , Dongmei Zhao 2
|
Background
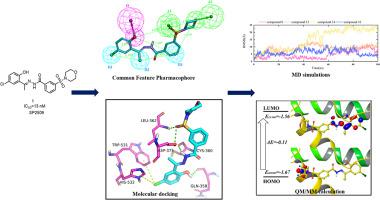
Histone lysine-specific demethylase 1 (LSD1) has become a potential anticancer target for the novel drug discovery. Recent reports have shown that SP2509 and its derivatives strongly inhibit LSD1 as allosteric inhibitors. However, the binding mechanism of these allosteric inhibitors in the allosteric site of LSD1 is not known yet.
Methods
The stability and binding mechanism of allosteric inhibitors in the binding site of LSD1 were evaluated by molecular docking, ligand-based pharmacophore, molecular dynamics (MD) simulations, molecular mechanics generalized born surface area (MM/GBSA) analysis, quantum mechanics/molecular mechanics (QM/MM) calculation and Hirshfeld surface analysis.
Results
The conformational geometry and the intermolecular interactions of allosteric inhibitors showed high binding affinity towards allosteric site of LSD1 with the neighboring amino acids (Gly358, Cys360, Leu362, Asp375 and Glu379). Meanwhile, MD simulations and MM/GBSA analysis were performed on selected allosteric inhibitors in complex with LSD1 protein, which confirmed the high stability and binding affinity of these inhibitors in the allosteric site of LSD1.
Conclusion
The simulation results revealed the crucial factors accounting for allosteric inhibitors of LSD1, including different protein–ligand interactions, the positions and conformations of key residues, and the ligands flexibilities. Meanwhile, a halogen bond interaction between chlorine atom of ligand and key residues Trp531 and His532 was recurrent in our analysis confirming its importance.
General significance
Overall, our research analyzed in depth the binding modes of allosteric inhibitors with LSD1 and could provide useful information for the design of novel allosteric inhibitors.
中文翻译:

对组蛋白赖氨酸特异性脱甲基酶 1 变构抑制的分子间洞察
背景
组蛋白赖氨酸特异性去甲基化酶 1 (LSD1) 已成为新药发现的潜在抗癌靶点。最近的报道表明,SP2509 及其衍生物作为变构抑制剂强烈抑制 LSD1。然而,这些变构抑制剂在 LSD1 变构位点的结合机制尚不清楚。
方法
通过分子对接、基于配体的药效团、分子动力学 (MD) 模拟、分子力学广义出生表面积 (MM/GBSA) 分析、量子力学/分子力学评估变构抑制剂在 LSD1 结合位点的稳定性和结合机制(QM/MM) 计算和 Hirshfeld 表面分析。
结果
变构抑制剂的构象几何学和分子间相互作用显示出对 LSD1 变构位点与相邻氨基酸(Gly358、Cys360、Leu362、Asp375 和 Glu379)的高结合亲和力。同时,对选定的与 LSD1 蛋白复合的变构抑制剂进行了 MD 模拟和 MM/GBSA 分析,证实了这些抑制剂在 LSD1 变构位点的高稳定性和结合亲和力。
结论
模拟结果揭示了导致 LSD1 变构抑制剂的关键因素,包括不同的蛋白质-配体相互作用、关键残基的位置和构象以及配体的灵活性。同时,配体的氯原子与关键残基 Trp531 和 His532 之间的卤键相互作用在我们的分析中反复出现,证实了其重要性。
一般意义
总的来说,我们的研究深入分析了变构抑制剂与 LSD1 的结合模式,可以为新型变构抑制剂的设计提供有用的信息。










































 京公网安备 11010802027423号
京公网安备 11010802027423号