当前位置:
X-MOL 学术
›
Angew. Chem. Int. Ed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of RNA Base Pairs and Complete Assignment of Nucleobase Resonances by Proton-Detected Solid-State NMR Spectroscopy at 100 kHz MAS
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2021-08-11 , DOI: 10.1002/anie.202107263 Philipp Innig Aguion 1 , John Kirkpatrick 1, 2 , Teresa Carlomagno 1, 2 , Alexander Marchanka 1
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2021-08-11 , DOI: 10.1002/anie.202107263 Philipp Innig Aguion 1 , John Kirkpatrick 1, 2 , Teresa Carlomagno 1, 2 , Alexander Marchanka 1
Affiliation
|
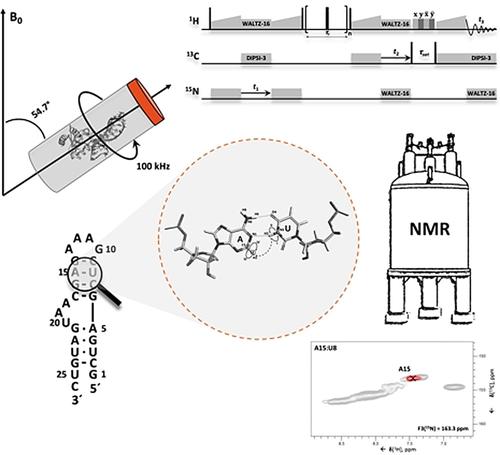
Knowledge of RNA structure, either in isolation or in complex, is fundamental to understand the mechanism of cellular processes. Solid-state NMR (ssNMR) is applicable to high molecular-weight complexes and does not require crystallization; thus, it is well-suited to study RNA as part of large multicomponent assemblies. Recently, we solved the first structures of both RNA and an RNA-protein complex by ssNMR using conventional 13C- and 15N-detection. This approach is limited by the severe overlap of the RNA peaks together with the low sensitivity of multidimensional experiments. Here, we overcome the limitations in sensitivity and resolution by using 1H-detection at fast MAS rates. We develop experiments that allow the identification of complete nucleobase spin-systems together with their site-specific base pair pattern using sub-milligram quantities of one uniformly labelled RNA sample. These experiments provide rapid access to RNA secondary structure by ssNMR in protein-RNA complexes of any size.
中文翻译:

通过 100 kHz MAS 质子检测固态核磁共振光谱鉴定 RNA 碱基对和核碱基共振的完整分配
RNA 结构的知识,无论是孤立的还是复杂的,是理解细胞过程机制的基础。固态核磁共振(ssNMR)适用于高分子量配合物,不需要结晶;因此,它非常适合作为大型多组件组件的一部分来研究 RNA。最近,我们使用常规13 C 和15 N 检测通过 ssNMR 解决了 RNA 和 RNA-蛋白质复合物的第一个结构。这种方法受限于 RNA 峰的严重重叠以及多维实验的低灵敏度。在这里,我们通过使用1快速 MAS 速率下的 H 检测。我们开发的实验允许使用亚毫克量的一个均匀标记的 RNA 样本来鉴定完整的核碱基自旋系统及其位点特异性碱基对模式。这些实验通过 ssNMR 在任何大小的蛋白质-RNA 复合物中提供了对 RNA 二级结构的快速访问。
更新日期:2021-10-19
中文翻译:

通过 100 kHz MAS 质子检测固态核磁共振光谱鉴定 RNA 碱基对和核碱基共振的完整分配
RNA 结构的知识,无论是孤立的还是复杂的,是理解细胞过程机制的基础。固态核磁共振(ssNMR)适用于高分子量配合物,不需要结晶;因此,它非常适合作为大型多组件组件的一部分来研究 RNA。最近,我们使用常规13 C 和15 N 检测通过 ssNMR 解决了 RNA 和 RNA-蛋白质复合物的第一个结构。这种方法受限于 RNA 峰的严重重叠以及多维实验的低灵敏度。在这里,我们通过使用1快速 MAS 速率下的 H 检测。我们开发的实验允许使用亚毫克量的一个均匀标记的 RNA 样本来鉴定完整的核碱基自旋系统及其位点特异性碱基对模式。这些实验通过 ssNMR 在任何大小的蛋白质-RNA 复合物中提供了对 RNA 二级结构的快速访问。











































 京公网安备 11010802027423号
京公网安备 11010802027423号