Current Proteomics ( IF 0.5 ) Pub Date : 2021-05-31 , DOI: 10.2174/1570164617999200720170153 Sangeeta Yadav 1 , Gautam Anand 1 , Vinay K Singh 2 , Dinesh Yadav 1
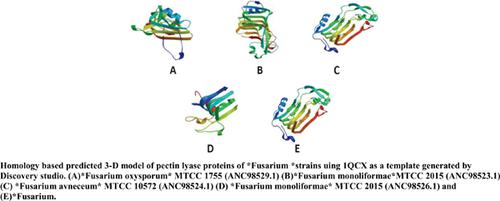
|
Aim: Molecular cloning and analysis of Pectin Iyase (PNL) genes from different strains of Fusarium for structural predictions and docking studies.
Background: PNLs cleave pectin by β-elimination resulting in the formation of 4,5-unsaturated oligogalacturonates, without affecting the ester content of the polymer chain and hence maintaining the specific aroma of fruits. Several PNL lyase genes from Aspergillus and Penicillium have been cloned, but the molecular biology of that from Fusarium has not been explored.
Objective: To obtain an insight into the three-dimensional structure of PNL of Fusarium.
Methods: PCR amplification-based molecular cloning of PNL genes from Fusarium strains, sequencing, and sequence analysis using bioinformatics tools for homology search, multiple sequence alignment, motif search, physiochemical characterization, phylogenetic tree construction, 3D structure prediction, and molecular docking were conducted.
Results: Five PNL genes were cloned from F. oxysporum MTCC1755, F. monoliforme var. subglutinans MTCC2015, F. avenaceum MTCC10572, and F. solani MTCC3004 using the PCR approach. Many conserved amino acids were found at several positions in all the PNL proteins. Phylogenetic analysis of these proteins with other pectinases revealed two major clusters representing members of lyases and hydrolases. In-silico characterization revealed stable PNL proteins. PNL proteins from different Fusarium strains were similar in structural features and biochemical properties owing to their similar primary sequence. Docking studies revealed that electrostatic forces and van der Waal and hydrogen bonds effectuate the interaction between the ligand and the enzyme. Aspartate, tyrosine, and tryptophan residues in the active site of the enzyme are responsible for ligand binding.
Conclusion: PNL from different Fusarium species show similarity at structural as well as biochemical level. PNL protein from F. moniliforme and F. solani was similar in properties except for the variation of single amino acid. Docking studies on the enzyme and different ligands provided an insight into the interacting residues and forces as well as the suitability of the substrate for catalysis.
中文翻译:

不同镰刀菌菌株果胶裂解酶蛋白的分子克隆和结构分析
目的:对不同镰刀菌菌株的果胶裂解酶 (PNL) 基因进行分子克隆和分析,用于结构预测和对接研究。
背景:PNLs 通过β-消除裂解果胶,形成4,5-不饱和低聚半乳糖醛酸酯,不影响聚合物链的酯含量,从而保持水果的特殊香气。已经克隆了来自曲霉属和青霉属的几种 PNL 裂解酶基因,但尚未探索来自镰刀菌属的 PNL 裂解酶基因。
目的:深入了解镰刀菌 PNL 的三维结构。
方法:对镰刀菌属菌株的 PNL 基因进行基于 PCR 扩增的分子克隆、测序和序列分析,使用生物信息学工具进行同源性搜索、多序列比对、基序搜索、理化表征、系统发育树构建、3D 结构预测和分子对接.
结果:从尖孢镰刀菌 MTCC1755、F. monoliforme var. 中克隆了五个 PNL 基因。谷蛋白 MTCC2015、F. avenaceum MTCC10572 和 F. solani MTCC3004 使用 PCR 方法。在所有 PNL 蛋白的几个位置发现了许多保守的氨基酸。这些蛋白质与其他果胶酶的系统发育分析揭示了代表裂解酶和水解酶成员的两个主要簇。计算机表征揭示了稳定的 PNL 蛋白。来自不同镰刀菌菌株的 PNL 蛋白由于其相似的一级序列而在结构特征和生化特性上相似。对接研究表明静电力和范德华力和氢键实现了配体和酶之间的相互作用。天冬氨酸、酪氨酸、
结论:来自不同镰刀菌属的 PNL 在结构和生化水平上表现出相似性。F. moniliforme 和 F. solani 的 PNL 蛋白除了单个氨基酸的变异外,在性质上相似。对酶和不同配体的对接研究提供了对相互作用残基和力以及底物催化适用性的深入了解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号