当前位置:
X-MOL 学术
›
FEBS Open Bio
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
JWES: a new pipeline for whole genome/exome sequence data processing, management, and gene-variant discovery, annotation, prediction, and genotyping
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-08-09 , DOI: 10.1002/2211-5463.13261 Zeeshan Ahmed 1, 2 , Eduard Gibert Renart 1 , Deepshikha Mishra 1 , Saman Zeeshan 3
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-08-09 , DOI: 10.1002/2211-5463.13261 Zeeshan Ahmed 1, 2 , Eduard Gibert Renart 1 , Deepshikha Mishra 1 , Saman Zeeshan 3
Affiliation
|
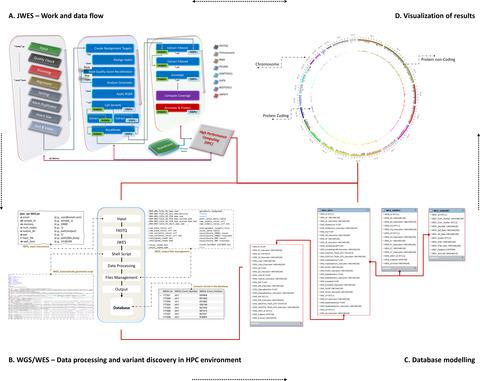
Whole genome and exome sequencing (WGS/WES) are the most popular next-generation sequencing (NGS) methodologies and are at present often used to detect rare and common genetic variants of clinical significance. We emphasize that automated sequence data processing, management, and visualization should be an indispensable component of modern WGS and WES data analysis for sequence assembly, variant detection (SNPs, SVs), imputation, and resolution of haplotypes. In this manuscript, we present a newly developed findable, accessible, interoperable, and reusable (FAIR) bioinformatics-genomics pipeline Java based Whole Genome/Exome Sequence Data Processing Pipeline (JWES) for efficient variant discovery and interpretation, and big data modeling and visualization. JWES is a cross-platform, user-friendly, product line application, that entails three modules: (a) data processing, (b) storage, and (c) visualization. The data processing module performs a series of different tasks for variant calling, the data storage module efficiently manages high-volume gene-variant data, and the data visualization module supports variant data interpretation with Circos graphs. The performance of JWES was tested and validated in-house with different experiments, using Microsoft Windows, macOS Big Sur, and UNIX operating systems. JWES is an open-source and freely available pipeline, allowing scientists to take full advantage of all the computing resources available, without requiring much computer science knowledge. We have successfully applied JWES for processing, management, and gene-variant discovery, annotation, prediction, and genotyping of WGS and WES data to analyze variable complex disorders. In summary, we report the performance of JWES with some reproducible case studies, using open access and in-house generated, high-quality datasets.
中文翻译:

JWES:全基因组/外显子组序列数据处理、管理和基因变异发现、注释、预测和基因分型的新管道
全基因组和外显子组测序 (WGS/WES) 是最流行的下一代测序 (NGS) 方法,目前常用于检测具有临床意义的罕见和常见遗传变异。我们强调,自动化序列数据处理、管理和可视化应该是现代 WGS 和 WES 数据分析中不可或缺的组成部分,用于序列组装、变异检测(SNP、SV)、插补和单倍型解析。在这份手稿中,我们展示了一个新开发的可查找、可访问、可互操作和可重用 (FAIR) 生物信息学-基因组学管道,基于 Java 的全基因组/外显子组序列数据处理管道 (JWES),用于高效的变异发现和解释,以及大数据建模和可视化. JWES 是一个跨平台、用户友好的产品线应用程序,这需要三个模块:(a) 数据处理、(b) 存储和 (c) 可视化。数据处理模块执行一系列不同的变异调用任务,数据存储模块高效管理大量基因变异数据,数据可视化模块支持使用 Circos 图表解释变异数据。JWES 的性能在内部通过不同的实验进行了测试和验证,使用 Microsoft Windows、macOS Big Sur 和 UNIX 操作系统。JWES 是一个开源和免费可用的管道,允许科学家充分利用所有可用的计算资源,而不需要太多的计算机科学知识。我们已成功将 JWES 应用于处理、管理和基因变异发现、注释、预测、和 WGS 和 WES 数据的基因分型,以分析可变的复杂疾病。总之,我们使用开放访问和内部生成的高质量数据集,通过一些可重复的案例研究报告了 JWES 的性能。
更新日期:2021-09-01
中文翻译:

JWES:全基因组/外显子组序列数据处理、管理和基因变异发现、注释、预测和基因分型的新管道
全基因组和外显子组测序 (WGS/WES) 是最流行的下一代测序 (NGS) 方法,目前常用于检测具有临床意义的罕见和常见遗传变异。我们强调,自动化序列数据处理、管理和可视化应该是现代 WGS 和 WES 数据分析中不可或缺的组成部分,用于序列组装、变异检测(SNP、SV)、插补和单倍型解析。在这份手稿中,我们展示了一个新开发的可查找、可访问、可互操作和可重用 (FAIR) 生物信息学-基因组学管道,基于 Java 的全基因组/外显子组序列数据处理管道 (JWES),用于高效的变异发现和解释,以及大数据建模和可视化. JWES 是一个跨平台、用户友好的产品线应用程序,这需要三个模块:(a) 数据处理、(b) 存储和 (c) 可视化。数据处理模块执行一系列不同的变异调用任务,数据存储模块高效管理大量基因变异数据,数据可视化模块支持使用 Circos 图表解释变异数据。JWES 的性能在内部通过不同的实验进行了测试和验证,使用 Microsoft Windows、macOS Big Sur 和 UNIX 操作系统。JWES 是一个开源和免费可用的管道,允许科学家充分利用所有可用的计算资源,而不需要太多的计算机科学知识。我们已成功将 JWES 应用于处理、管理和基因变异发现、注释、预测、和 WGS 和 WES 数据的基因分型,以分析可变的复杂疾病。总之,我们使用开放访问和内部生成的高质量数据集,通过一些可重复的案例研究报告了 JWES 的性能。











































 京公网安备 11010802027423号
京公网安备 11010802027423号