Comparative Biochemistry and Physiology B: Biochemistry & Molecular Biology ( IF 1.9 ) Pub Date : 2021-08-06 , DOI: 10.1016/j.cbpb.2021.110662 Daichi Yano 1 , Kouji Uda 1 , Masakazu Nara 2 , Tomohiko Suzuki 1
|
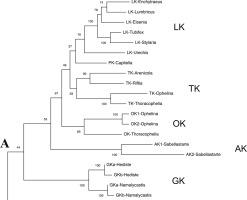
Opheline kinase (OK) is one of the phosphagen kinases (PKs) restricted to annelids, but the amino acid sequence has not been determined yet. The OK enzyme was isolated in 1966 from the polychaete Ophelia neglecta (Opheliidae) and shown to have somewhat broader activities for the various substrates opheline, lombricine and taurocyamine. To determine the OK sequence, we analyzed the RNA sequencing data for Ophelina sp. and Thoracophelia sp., belonging to Opheliidae. Four PK sequences, namely, taurocyamine kinase (TK), creatine kinase (CK), mitochondrial CK (MiCK) and putative OK, were identified in both species, and the recombinant Ophelina enzymes were expressed in E. coli and purified. Since the substrate opheline was not commercially available, we used the partial activity toward taurocyamine to infer the enzyme specificity. The putative Ophelina OK showed lower activity to taurocyamine with a Vmax/Km nearly identical to a previously published value for an OK from a related species Ophelia neglecta. Under the same conditions, the true Ophelina TK showed much higher activity. Thus, the putative Ophelina enzyme was determined to be OK. The amino acid sequence alignment indicated that Ophelina and Thoracophelia OKs have five amino acid deletions in the GS region, like those of LKs and AKs, and the guanidino substrate specific residue was Lys, the same as LKs. In the phylogenetic tree constructed from annelid PK amino acid sequences, the OK sequences formed a distinct cluster, and it was placed near the TK and lombricine kinase (LK) clusters. This is the first report of the amino acid sequence for the OK enzyme.
中文翻译:

环节动物中磷酸原激酶的多样性:关于假定的蛇毒激酶的第一份序列报告
Opheline 激酶 (OK) 是一种仅限于环节动物的磷酸原激酶 (PK),但其氨基酸序列尚未确定。OK 酶于 1966 年从多毛类Ophelia ignorea (Opheliidae)中分离出来,并显示出对各种底物 Opheline、lombricine 和 taurocyamine 具有更广泛的活性。为了确定 OK 序列,我们分析了Ophelina sp的 RNA 测序数据。和Thoracophelia sp.,属于 Opheliidae。在两个物种中鉴定出4个PK序列,即牛磺胺激酶(TK)、肌酸激酶(CK)、线粒体CK(MiCK)和推定的OK,重组Ophelina酶在大肠杆菌中表达和净化。由于底物 Opheline 不是市售的,我们使用对牛磺氰胺的部分活性来推断酶特异性。推定的Ophelina OK 对牛磺氰胺的活性较低,其 V max /K m与先前公布的相关物种Ophelia ignorea 的 OK 值几乎相同。在相同条件下,真正的 Ophelina TK 表现出更高的活性。因此,推定的Ophelina酶被确定为 OK。氨基酸序列比对表明Ophelina和ThoracopheliaOKs在GS区有5个氨基酸缺失,与LKs和AKs相同,胍基底物特异性残基为Lys,与LKs相同。在环节动物PK氨基酸序列构建的系统发育树中,OK序列形成一个独特的簇,位于TK和Lombricine激酶(LK)簇附近。这是OK酶氨基酸序列的首次报道。











































 京公网安备 11010802027423号
京公网安备 11010802027423号