Metabolic Engineering ( IF 6.8 ) Pub Date : 2021-07-31 , DOI: 10.1016/j.ymben.2021.07.014 Ekaterina Kozaeva 1 , Svetlana Volkova 1 , Marta R A Matos 1 , Mariela P Mezzina 1 , Tune Wulff 1 , Daniel C Volke 1 , Lars K Nielsen 2 , Pablo I Nikel 1
|
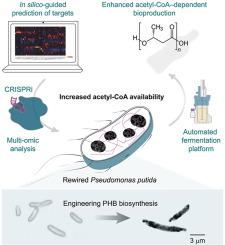
Pseudomonas putida is evolutionarily endowed with features relevant for bioproduction, especially under harsh operating conditions. The rich metabolic versatility of this species, however, comes at the price of limited formation of acetyl-coenzyme A (CoA) from sugar substrates. Since acetyl-CoA is a key metabolic precursor for a number of added-value products, in this work we deployed an in silico-guided rewiring program of central carbon metabolism for upgrading P. putida as a host for acetyl-CoA–dependent bioproduction. An updated kinetic model, integrating fluxomics and metabolomics datasets in addition to manually-curated information of enzyme mechanisms, identified targets that would lead to increased acetyl-CoA levels. Based on these predictions, a set of plasmids based on clustered regularly interspaced short palindromic repeats (CRISPR) and dead CRISPR-associated protein 9 (dCas9) was constructed to silence genes by CRISPR interference (CRISPRi). Dynamic reduction of gene expression of two key targets (gltA, encoding citrate synthase, and the essential accA gene, encoding subunit A of the acetyl-CoA carboxylase complex) mediated an 8-fold increase in the acetyl-CoA content of rewired P. putida. Poly(3-hydroxybutyrate) (PHB) was adopted as a proxy of acetyl-CoA availability, and two synthetic pathways were engineered for biopolymer accumulation. By including cell morphology as an extra target for the CRISPRi approach, fully rewired P. putida strains programmed for PHB accumulation had a 5-fold increase in PHB titers in bioreactor cultures using glucose. Thus, the strategy described herein allowed for rationally redirecting metabolic fluxes in P. putida from central metabolism towards product biosynthesis—especially relevant when deletion of essential pathways is not an option.
中文翻译:

基本代谢节点的模型引导动态控制可促进重新连接的恶臭假单胞菌中依赖乙酰辅酶 A 的生物生产
恶臭假单胞菌在进化上具有与生物生产相关的特征,尤其是在恶劣的操作条件下。然而,该物种丰富的代谢多样性是以糖底物有限形成乙酰辅酶 A (CoA) 为代价的。由于乙酰辅酶 A 是许多附加值产品的关键代谢前体,在这项工作中,我们部署了一个计算机引导的中央碳代谢重新布线程序,用于升级恶臭假单胞菌作为乙酰辅酶A依赖性生物生产的宿主。更新的动力学模型将通量组学和代谢组学数据集以及酶机制的手动管理信息整合在一起,确定了会导致乙酰辅酶 A 水平增加的目标。基于这些预测,构建了一组基于成簇的规则间隔短回文重复序列 (CRISPR) 和死 CRISPR 相关蛋白 9 (dCas9) 的质粒,以通过 CRISPR 干扰 (CRISPRi) 沉默基因。两个关键靶标(gltA、柠檬酸合酶编码和必需的ACCA基因,编码乙酰辅酶 A 羧化酶复合物的亚基 A)的基因表达的动态减少介导重新接线的P的乙酰辅酶 A 含量增加 8 倍。恶臭. 聚(3-羟基丁酸)(PHB)被用作乙酰辅酶A可用性的代表,并且设计了两种合成途径用于生物聚合物的积累。通过将细胞形态作为 CRISPRi 方法的一个额外目标,完全重新连接的恶臭假单胞菌菌株在使用葡萄糖的生物反应器培养物中的 PHB 滴度增加了 5 倍。因此,中描述的策略允许合理地将恶臭假单胞菌中的代谢通量从中心代谢转向产物生物合成——尤其是在不能选择删除必需途径时。











































 京公网安备 11010802027423号
京公网安备 11010802027423号