Structure ( IF 4.4 ) Pub Date : 2021-07-30 , DOI: 10.1016/j.str.2021.07.001 Nicholas J Fowler 1 , Adnan Sljoka 2 , Mike P Williamson 1
|
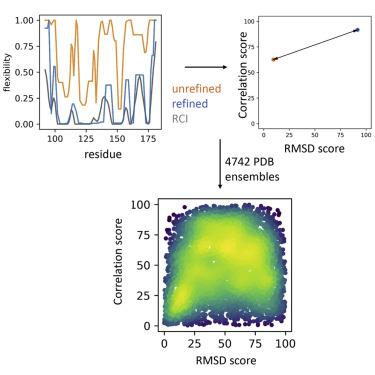
The program ANSURR measures the accuracy of NMR structures by comparing rigidity obtained from experimental backbone chemical shifts and from structures. We report on ANSURR analysis of 7,000 PDB NMR ensembles within the Protein Data Bank, which can be found at ansurr.com. The accuracy of NMR structures progressively improved up until 2005, but since then, it has plateaued. Most structures have accurate secondary structure, but are generally too floppy, particularly in loops. Thus, there is a need for more experimental restraints in loops. Currently, the best predictors of accuracy are Ramachandran distribution and the number of NOE restraints per residue. The precision of structures within the ensemble correlates well with accuracy, as does the number of hydrogen bond restraints per residue. Structure accuracy is improved when other components (such as additional polypeptide chains or ligands) are included.
中文翻译:

蛋白质数据库中 NMR 蛋白质结构的准确性
ANSURR 程序通过比较从实验骨架化学位移和结构获得的刚度来测量 NMR 结构的准确性。我们报告了蛋白质数据库中 7,000 个 PDB NMR 集合的 ANSURR 分析,该数据库可在 ansurr.com 上找到。直到 2005 年,NMR 结构的准确性逐渐提高,但从那时起,它就趋于平稳。大多数结构具有精确的二级结构,但通常过于松散,特别是在循环中。因此,需要在循环中进行更多的实验限制。目前,准确度的最佳预测指标是 Ramachandran 分布和每个残基的 NOE 限制数。整体内结构的精度与精度密切相关,每个残基的氢键限制数量也是如此。











































 京公网安备 11010802027423号
京公网安备 11010802027423号