当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Improving Measures of Chemical Structural Similarity Using Machine Learning on Chemical–Genetic Interactions
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-07-28 , DOI: 10.1021/acs.jcim.0c00993 Hamid Safizadeh 1, 2 , Scott W Simpkins 3 , Justin Nelson 3 , Sheena C Li 4, 5 , Jeff S Piotrowski 5 , Mami Yoshimura 5 , Yoko Yashiroda 5 , Hiroyuki Hirano 5 , Hiroyuki Osada 5 , Minoru Yoshida 5, 6 , Charles Boone 4, 5, 7 , Chad L Myers 2, 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-07-28 , DOI: 10.1021/acs.jcim.0c00993 Hamid Safizadeh 1, 2 , Scott W Simpkins 3 , Justin Nelson 3 , Sheena C Li 4, 5 , Jeff S Piotrowski 5 , Mami Yoshimura 5 , Yoko Yashiroda 5 , Hiroyuki Hirano 5 , Hiroyuki Osada 5 , Minoru Yoshida 5, 6 , Charles Boone 4, 5, 7 , Chad L Myers 2, 3
Affiliation
|
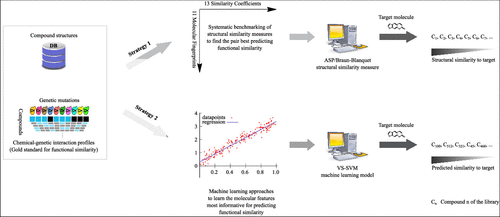
A common strategy for identifying molecules likely to possess a desired biological activity is to search large databases of compounds for high structural similarity to a query molecule that demonstrates this activity, under the assumption that structural similarity is predictive of similar biological activity. However, efforts to systematically benchmark the diverse array of available molecular fingerprints and similarity coefficients have been limited by a lack of large-scale datasets that reflect biological similarities of compounds. To elucidate the relative performance of these alternatives, we systematically benchmarked 11 different molecular fingerprint encodings, each combined with 13 different similarity coefficients, using a large set of chemical–genetic interaction data from the yeast Saccharomyces cerevisiae as a systematic proxy for biological activity. We found that the performance of different molecular fingerprints and similarity coefficients varied substantially and that the all-shortest path fingerprints paired with the Braun-Blanquet similarity coefficient provided superior performance that was robust across several compound collections. We further proposed a machine learning pipeline based on support vector machines that offered a fivefold improvement relative to the best unsupervised approach. Our results generally suggest that using high-dimensional chemical–genetic data as a basis for refining molecular fingerprints can be a powerful approach for improving prediction of biological functions from chemical structures.
中文翻译:

使用机器学习改进化学-遗传相互作用的化学结构相似性测量
识别可能具有所需生物活性的分子的常见策略是在结构相似性预示着相似的生物活性的假设下,搜索大型化合物数据库以寻找与证明该活性的查询分子的高度结构相似性。然而,由于缺乏反映化合物生物相似性的大规模数据集,系统地对各种可用分子指纹和相似系数进行基准测试的努力受到了限制。为了阐明这些替代品的相对性能,我们系统地对 11 种不同的分子指纹编码进行了基准测试,每种编码都结合了 13 种不同的相似系数,使用来自酵母酿酒酵母的大量化学-遗传相互作用数据作为生物活性的系统代理。我们发现不同分子指纹和相似系数的性能差异很大,并且与 Braun-Blanquet 相似系数配对的全最短路径指纹提供了卓越的性能,在多个化合物集合中表现稳健。我们进一步提出了一种基于支持向量机的机器学习管道,相对于最佳无监督方法提供了五倍的改进。我们的结果通常表明,使用高维化学遗传数据作为提炼分子指纹的基础可以成为改善化学结构生物功能预测的有效方法。
更新日期:2021-09-27
中文翻译:

使用机器学习改进化学-遗传相互作用的化学结构相似性测量
识别可能具有所需生物活性的分子的常见策略是在结构相似性预示着相似的生物活性的假设下,搜索大型化合物数据库以寻找与证明该活性的查询分子的高度结构相似性。然而,由于缺乏反映化合物生物相似性的大规模数据集,系统地对各种可用分子指纹和相似系数进行基准测试的努力受到了限制。为了阐明这些替代品的相对性能,我们系统地对 11 种不同的分子指纹编码进行了基准测试,每种编码都结合了 13 种不同的相似系数,使用来自酵母酿酒酵母的大量化学-遗传相互作用数据作为生物活性的系统代理。我们发现不同分子指纹和相似系数的性能差异很大,并且与 Braun-Blanquet 相似系数配对的全最短路径指纹提供了卓越的性能,在多个化合物集合中表现稳健。我们进一步提出了一种基于支持向量机的机器学习管道,相对于最佳无监督方法提供了五倍的改进。我们的结果通常表明,使用高维化学遗传数据作为提炼分子指纹的基础可以成为改善化学结构生物功能预测的有效方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号