Journal of Structural Biology ( IF 3.0 ) Pub Date : 2021-07-25 , DOI: 10.1016/j.jsb.2021.107773 Vitor Medeiros Almeida 1 , Apala Chaudhuri 2 , Marcus Vinicius Cangussu Cardoso 3 , Bruno Yasui Matsuyama 1 , Gláucio Monteiro Ferreira 4 , Gustavo Henrique Goulart Trossini 4 , Roberto Kopke Salinas 1 , J Patrick Loria 5 , Sandro Roberto Marana 1
|
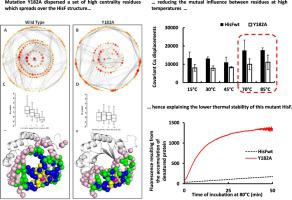
Centralities determined from Residue Interaction Networks (RIN) in proteins have been used to predict aspects of their structure and dynamics. Here, we correlate the Eigenvector Centrality (Ec) with the rate constant for thermal denaturation (kden) of the HisF protein from Thermotoga maritima based on 12 single alanine substitution mutants. The molecular basis for this correlation was further explored by studying a mutant containing a replacement of a high Ec residue, Y182A, which displayed increased kden at 80 °C. The crystallographic structure of this mutant showed few changes, mostly in two flexible loops. The 1H-15N -HSQC showed only subtle changes of cross peak positions for residues located near the mutation site and scattered throughout the structure. However, the comparison of the RIN showed that Y182 is the vertex of a set of high centrality residues that spreads throughout the HisF structure, which is lacking in the mutant. Cross-correlation displacements of Cα calculated from a molecular dynamics simulation at different temperatures showed that the Y182A mutation reduced the correlated movements in the HisF structure above 70 °C. 1H-15N NMR chemical shift covariance using temperature as perturbation were consistent with these results. In conclusion the increase in temperature drives the structure of the mutant HisF-Y182A into a less connected state, richer in non-concerted motions, located predominantly in the C-terminal half of the protein where Y182 is placed. Conversely, wild-type HisF responds to increased temperature as a single unit. Hence the replacement of a high Ec residue alters the distribution of thermal energy through HisF structure.
中文翻译:

高中心性残基在蛋白质动力学和热稳定性中的作用
根据蛋白质中的残基相互作用网络 (RIN) 确定的中心性已用于预测其结构和动力学的各个方面。在这里,我们基于 12 个单丙氨酸取代突变体,将特征向量中心性 (E c ) 与海栖热袍菌HisF 蛋白的热变性速率常数 ( k den ) 相关联。通过研究含有高 E c残基替换的突变体 Y182A,进一步探索了这种相关性的分子基础,该突变体在 80 °C 下表现出增加的k den 。该突变体的晶体结构几乎没有变化,主要是两个柔性环。 1 H- 15 N -HSQC 仅显示位于突变位点附近并分散在整个结构中的残基交叉峰位置的细微变化。然而,RIN 的比较表明,Y182 是一组高中心性残基的顶点,这些残基遍布整个 HisF 结构,这是突变体中所缺乏的。根据不同温度下的分子动力学模拟计算出的 Cα 互相关位移表明,Y182A 突变减少了 HisF 结构在 70 °C 以上的相关运动。使用温度作为扰动的1 H- 15 N NMR化学位移协方差与这些结果一致。总之,温度的升高驱动突变体 HisF-Y182A 的结构进入连接较少的状态,非协同运动更丰富,主要位于 Y182 所在的蛋白质 C 端一半。相反,野生型 HisF 作为一个单位对温度升高做出反应。 因此,高 E c残基的替换改变了通过 HisF 结构的热能分布。











































 京公网安备 11010802027423号
京公网安备 11010802027423号