Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrated Addressable Dynamic Droplet Array (aDDA) as Sub-Nanoliter Reactors for High-Coverage Genome Sequencing of Single Yeast Cells
Small ( IF 13.0 ) Pub Date : 2021-07-23 , DOI: 10.1002/smll.202100325 Chunyu Li 1 , Yanhai Gong 1 , Xixian Wang 1 , Jian Xu 1 , Bo Ma 1
Small ( IF 13.0 ) Pub Date : 2021-07-23 , DOI: 10.1002/smll.202100325 Chunyu Li 1 , Yanhai Gong 1 , Xixian Wang 1 , Jian Xu 1 , Bo Ma 1
Affiliation
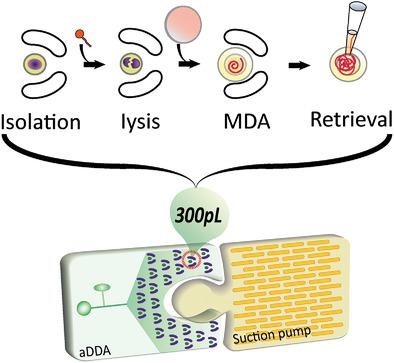
|
An addressable dynamic droplet array (aDDA) is presented that combines the advantages of static droplet arrays and continuous-flow droplet platforms. Modular fabrication is employed to create a self-contained integrated aDDA. All the sample preparation steps, including single-cell isolation, cell lysis, amplification, and product retrieval, are performed in sequence within a sub-nanoliter (≈300 pL) droplet. Sequencing-based validation suggests that aDDA reduces the amplification bias of multiple displacement amplification (MDA) and elevates the percentage of one-yeast-cell genome recovery to 91%, as compared to the average of 26% using conventional, 20 µL volume MDA reactions. Thus, aDDA is a valuable addition to the toolbox for high-genome-coverage sequencing of single microbial cells.
中文翻译:

集成可寻址动态液滴阵列 (aDDA) 作为亚纳升反应器,用于单酵母细胞的高覆盖基因组测序
提出了一种可寻址的动态液滴阵列 (aDDA),它结合了静态液滴阵列和连续流动液滴平台的优点。采用模块化制造来创建独立的集成 aDDA。所有样品制备步骤,包括单细胞分离、细胞裂解、扩增和产物回收,均在亚纳升 (≈300 pL) 液滴内依次执行。基于测序的验证表明,与使用常规 20 µL 体积 MDA 反应的平均值 26% 相比,aDDA 降低了多重置换扩增 (MDA) 的扩增偏差,并将单酵母细胞基因组回收率提高到 91% . 因此,aDDA 是对单个微生物细胞的高基因组覆盖测序工具箱的宝贵补充。
更新日期:2021-09-16
中文翻译:

集成可寻址动态液滴阵列 (aDDA) 作为亚纳升反应器,用于单酵母细胞的高覆盖基因组测序
提出了一种可寻址的动态液滴阵列 (aDDA),它结合了静态液滴阵列和连续流动液滴平台的优点。采用模块化制造来创建独立的集成 aDDA。所有样品制备步骤,包括单细胞分离、细胞裂解、扩增和产物回收,均在亚纳升 (≈300 pL) 液滴内依次执行。基于测序的验证表明,与使用常规 20 µL 体积 MDA 反应的平均值 26% 相比,aDDA 降低了多重置换扩增 (MDA) 的扩增偏差,并将单酵母细胞基因组回收率提高到 91% . 因此,aDDA 是对单个微生物细胞的高基因组覆盖测序工具箱的宝贵补充。











































 京公网安备 11010802027423号
京公网安备 11010802027423号