当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Inducible Protein Degradation to Understand Genome Architecture
Biochemistry ( IF 2.9 ) Pub Date : 2021-07-22 , DOI: 10.1021/acs.biochem.1c00306 Alexi Tallan 1, 2 , Benjamin Z Stanton 1, 2, 3, 4
Biochemistry ( IF 2.9 ) Pub Date : 2021-07-22 , DOI: 10.1021/acs.biochem.1c00306 Alexi Tallan 1, 2 , Benjamin Z Stanton 1, 2, 3, 4
Affiliation
|
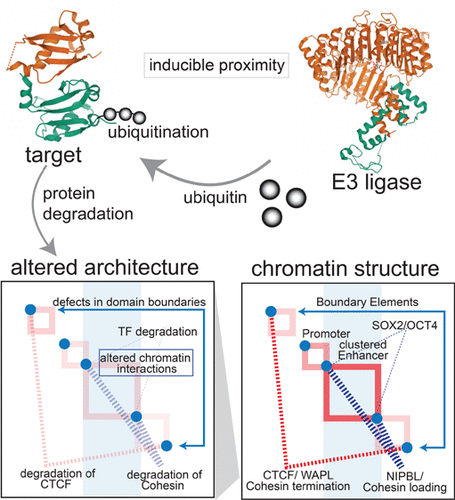
We review exciting recent advances in protein degradation, with a focus on chromatin structure. In our analysis of the literature, we highlight studies of kinetic control of protein stability for cohesin, condensin, ATP-dependent chromatin remodeling, and pioneer transcription factors. With new connections emerging between chromatin remodeling and genome structure, we anticipate exciting developments at the intersection of these topics to be revealed in the coming years. Moreover, we pay special attention to the 20-year anniversary of PROTACs, with an overview of E3 ligase/target pairings and central questions that might lead to the next generation of PROTACs with an expanded scope and generality. While steady-state experimental measurements with constitutive genome editing are impactful, we highlight complementary approaches for rapid kinetic protein degradation to uncover early targeting functions and to understand the central determinants of genome structure–function relationships.
中文翻译:

诱导蛋白降解以了解基因组结构
我们回顾了蛋白质降解方面令人兴奋的最新进展,重点是染色质结构。在我们对文献的分析中,我们重点研究了黏连蛋白、凝聚素、ATP 依赖性染色质重塑和先驱转录因子的蛋白质稳定性动力学控制研究。随着染色质重塑和基因组结构之间出现新的联系,我们预计这些主题的交叉点将在未来几年内出现令人兴奋的发展。此外,我们特别关注 PROTACs 20 周年,概述了 E3 连接酶/靶标配对和可能导致具有扩展范围和通用性的下一代 PROTACs 的核心问题。虽然本构基因组编辑的稳态实验测量具有影响力,
更新日期:2021-08-10
中文翻译:

诱导蛋白降解以了解基因组结构
我们回顾了蛋白质降解方面令人兴奋的最新进展,重点是染色质结构。在我们对文献的分析中,我们重点研究了黏连蛋白、凝聚素、ATP 依赖性染色质重塑和先驱转录因子的蛋白质稳定性动力学控制研究。随着染色质重塑和基因组结构之间出现新的联系,我们预计这些主题的交叉点将在未来几年内出现令人兴奋的发展。此外,我们特别关注 PROTACs 20 周年,概述了 E3 连接酶/靶标配对和可能导致具有扩展范围和通用性的下一代 PROTACs 的核心问题。虽然本构基因组编辑的稳态实验测量具有影响力,











































 京公网安备 11010802027423号
京公网安备 11010802027423号