当前位置:
X-MOL 学术
›
Environ. Sci. Technol. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Quantitative SARS-CoV-2 Alpha Variant B.1.1.7 Tracking in Wastewater by Allele-Specific RT-qPCR
Environmental Science & Technology Letters ( IF 8.9 ) Pub Date : 2021-07-18 , DOI: 10.1021/acs.estlett.1c00375 Wei Lin Lee 1, 2 , Maxim Imakaev 3 , Federica Armas 1, 2 , Kyle A. McElroy 3 , Xiaoqiong Gu 1, 2 , Claire Duvallet 3 , Franciscus Chandra 1, 2 , Hongjie Chen 1, 2 , Mats Leifels 4 , Samuel Mendola 3 , Róisín Floyd-O’Sullivan 3 , Morgan M. Powell 3 , Shane T. Wilson 3 , Karl L. J. Berge 1, 2 , Claire Y. J. Lim 1, 2 , Fuqing Wu 5, 6 , Amy Xiao 5, 6 , Katya Moniz 5, 6 , Newsha Ghaeli 3 , Mariana Matus 3 , Janelle Thompson 2, 4, 7 , Eric J. Alm 1, 2, 5, 6, 8
Environmental Science & Technology Letters ( IF 8.9 ) Pub Date : 2021-07-18 , DOI: 10.1021/acs.estlett.1c00375 Wei Lin Lee 1, 2 , Maxim Imakaev 3 , Federica Armas 1, 2 , Kyle A. McElroy 3 , Xiaoqiong Gu 1, 2 , Claire Duvallet 3 , Franciscus Chandra 1, 2 , Hongjie Chen 1, 2 , Mats Leifels 4 , Samuel Mendola 3 , Róisín Floyd-O’Sullivan 3 , Morgan M. Powell 3 , Shane T. Wilson 3 , Karl L. J. Berge 1, 2 , Claire Y. J. Lim 1, 2 , Fuqing Wu 5, 6 , Amy Xiao 5, 6 , Katya Moniz 5, 6 , Newsha Ghaeli 3 , Mariana Matus 3 , Janelle Thompson 2, 4, 7 , Eric J. Alm 1, 2, 5, 6, 8
Affiliation
|
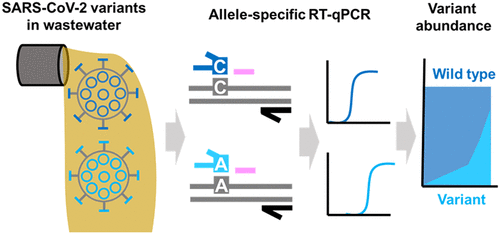
The critical need for surveillance of SARS-CoV-2 variants of concern has prompted the development of methods that can track variants in wastewater. Here, we develop and present an open-source method based on allele-specific RT-qPCR (AS RT-qPCR) that detects and quantifies the B.1.1.7 variant, targeting spike protein mutations at three independent genomic loci that are highly predictive of B.1.1.7 (HV69/70del, Y144del, and A570D). Our assays can reliably detect and quantify low levels of B.1.1.7 with low cross-reactivity, and at variant proportions down to 1% in a background of mixed SARS-CoV-2. Applying our method to wastewater samples from the United States, we track the occurrence of B.1.1.7 over time in 19 communities. AS RT-qPCR results align with clinical trends, and summation of B.1.1.7 and wild-type sequences quantified by our assays matches SARS-CoV-2 levels indicated by the U.S. CDC N1 and N2 assays. This work paves the way for AS RT-qPCR as a method for rapid inexpensive surveillance of SARS-CoV-2 variants in wastewater.
中文翻译:

通过等位基因特异性 RT-qPCR 对废水中的 SARS-CoV-2 Alpha 变体 B.1.1.7 进行定量跟踪
对关注的 SARS-CoV-2 变异体进行监测的迫切需求促使开发了可以跟踪废水中变异体的方法。在这里,我们开发并提出了一种基于等位基因特异性 RT-qPCR (AS RT-qPCR) 的开源方法,该方法检测和量化 B.1.1.7 变体,针对三个独立基因组位点的刺突蛋白突变,具有高度预测性B.1.1.7(HV69/70del、Y144del 和 A570D)。我们的检测方法可以可靠地检测和量化低水平的 B.1.1.7,具有低交叉反应性,并且在混合 SARS-CoV-2 背景下的变异比例低至 1%。将我们的方法应用于来自美国的废水样本,我们跟踪了 19 个社区随时间推移 B.1.1.7 的发生情况。AS RT-qPCR 结果与临床趋势以及 B.1.1 的总和一致。7 和野生型序列通过我们的检测定量与美国 CDC N1 和 N2 检测表明的 SARS-CoV-2 水平相匹配。这项工作为 AS RT-qPCR 作为一种快速廉价监测废水中 SARS-CoV-2 变体的方法铺平了道路。
更新日期:2021-08-10
中文翻译:

通过等位基因特异性 RT-qPCR 对废水中的 SARS-CoV-2 Alpha 变体 B.1.1.7 进行定量跟踪
对关注的 SARS-CoV-2 变异体进行监测的迫切需求促使开发了可以跟踪废水中变异体的方法。在这里,我们开发并提出了一种基于等位基因特异性 RT-qPCR (AS RT-qPCR) 的开源方法,该方法检测和量化 B.1.1.7 变体,针对三个独立基因组位点的刺突蛋白突变,具有高度预测性B.1.1.7(HV69/70del、Y144del 和 A570D)。我们的检测方法可以可靠地检测和量化低水平的 B.1.1.7,具有低交叉反应性,并且在混合 SARS-CoV-2 背景下的变异比例低至 1%。将我们的方法应用于来自美国的废水样本,我们跟踪了 19 个社区随时间推移 B.1.1.7 的发生情况。AS RT-qPCR 结果与临床趋势以及 B.1.1 的总和一致。7 和野生型序列通过我们的检测定量与美国 CDC N1 和 N2 检测表明的 SARS-CoV-2 水平相匹配。这项工作为 AS RT-qPCR 作为一种快速廉价监测废水中 SARS-CoV-2 变体的方法铺平了道路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号