Journal of Virological Methods ( IF 2.2 ) Pub Date : 2021-07-09 , DOI: 10.1016/j.jviromet.2021.114230 Mark Ciesielski 1 , Denene Blackwood 1 , Thomas Clerkin 1 , Raul Gonzalez 2 , Hannah Thompson 2 , Allison Larson 2 , Rachel Noble 1
|
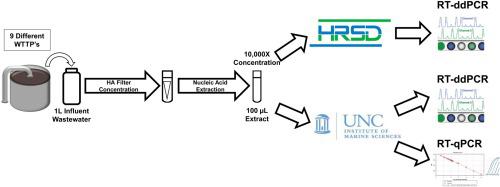
Throughout the COVID-19 global pandemic there has been significant interest and investment in using Wastewater-Based Epidemiology (WBE) for surveillance of viral pathogen presence and infections at the community level. There has been a push for widescale implementation of standardized protocols to quantify viral loads in a range of wastewater systems. To address concerns regarding sensitivity, limits of quantification, and large-scale reproducibility, a comparison of two similar workflows using RT-qPCR and RT-ddPCR was conducted. Sixty raw wastewater influent samples were acquired from nine distinct wastewater treatment plants (WWTP’s) served by the Hampton Roads Sanitation District (HRSD, Virginia Beach, Virginia) over a 6-month period beginning March 9th, 2020. Common reagents, controls, master mixes and nucleic acid extracts were shared between two individual processing groups based out of HRSD and the UNC Chapel Hill Institute of Marine Sciences (IMS, Morehead City, North Carolina). Samples were analyzed in parallel using One-Step RT-qPCR and One-Step RT-ddPCR with Nucleocapsid Protein 2 (N2) specific primers and probe. Influent SARS-CoV-2 N2 concentrations steadily increased over time spanning a range from non-detectable to 2.13E + 05 copies/L. Systematic dilution of the extracts indicated that inhibitory components in the wastewater matrices did not significantly impede the detection of a positive N2 signal for either workflow. The RT-ddPCR workflow had a greater analytical sensitivity with a lower Limit of Detection (LOD) at 0.066 copies/μl of template compared to RT-qPCR with a calculated LOD of 12.0 copies/μL of template. Interlaboratory comparisons using non-parametric correlation analysis demonstrated that there was a strong, significant, positive correlation between split extracts when employing RT-ddPCR for analysis with a ρ value of 0.86.
中文翻译:

评估 RT-ddPCR 和 RT-qPCR 定量废水中 SARS-CoV-2 的灵敏度和重现性
在 COVID-19 全球大流行期间,人们对使用基于废水的流行病学 (WBE) 来监测社区层面的病毒病原体存在和感染产生了浓厚的兴趣并进行了投资。人们一直在推动大规模实施标准化协议来量化一系列废水系统中的病毒载量。为了解决有关灵敏度、定量限制和大规模重现性的问题,对使用 RT-qPCR 和 RT-ddPCR 的两个类似工作流程进行了比较。自 2020 年 3 月 9 日开始的 6 个月内,从汉普顿路卫生区(弗吉尼亚州弗吉尼亚海滩 HRSD)服务的 9 个不同废水处理厂 (WWTP) 采集了 60 个原废水进水样品。 常用试剂、对照品、预混液核酸提取物在 HRSD 和北卡罗来纳大学教堂山海洋科学研究所(IMS,北卡罗来纳州莫尔黑德市)的两个单独处理小组之间共享。使用一步 RT-qPCR 和一步 RT-ddPCR 以及核衣壳蛋白 2 (N2) 特异性引物和探针对样品进行并行分析。流入的 SARS-CoV-2 N2 浓度随着时间的推移稳步增加,范围从不可检测到 2.13E + 05 拷贝/升。提取物的系统稀释表明废水基质中的抑制成分并没有显着妨碍任一工作流程中正 N2 信号的检测。与计算出的检测限 (LOD) 为 12.0 拷贝/μL 模板的 RT-qPCR 相比,RT-ddPCR 工作流程具有更高的分析灵敏度和更低的检测限 (LOD),为 0.066 拷贝/μL 模板。 使用非参数相关分析的实验室间比较表明,当采用 RT-ddPCR 进行分析时,分割提取物之间存在强烈、显着的正相关性,ρ 值为 0.86。











































 京公网安备 11010802027423号
京公网安备 11010802027423号