当前位置:
X-MOL 学术
›
FEBS Open Bio
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
EvoProDom: evolutionary modeling of protein families by assessing translocations of protein domains
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-07-01 , DOI: 10.1002/2211-5463.13245 Gon Carmi 1 , Alessandro Gorohovski 1 , Milana Frenkel-Morgenstern 1
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-07-01 , DOI: 10.1002/2211-5463.13245 Gon Carmi 1 , Alessandro Gorohovski 1 , Milana Frenkel-Morgenstern 1
Affiliation
|
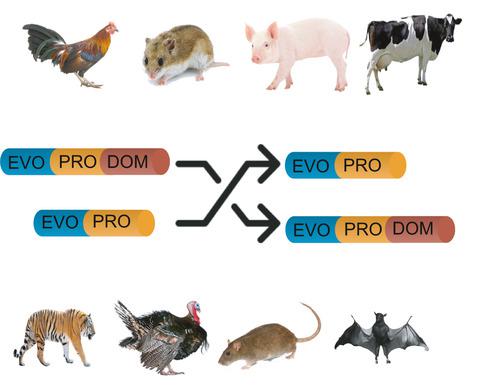
Here, we introduce a novel ‘evolution of protein domains’ (EvoProDom) model for describing the evolution of proteins based on the ‘mix and merge’ of protein domains. We assembled and integrated genomic and proteomic data comprising protein domain content and orthologous proteins from 109 organisms. In EvoProDom, we characterized evolutionary events, particularly, translocations, as reciprocal exchanges of protein domains between orthologous proteins in different organisms. We showed that protein domains that translocate with highly frequency are generated by transcripts enriched in trans-splicing events, that is, the generation of novel transcripts from the fusion of two distinct genes. In EvoProDom, we describe a general method to collate orthologous protein annotation from KEGG, and protein domain content from protein sequences using tools such as KoFamKOAL and Pfam. To summarize, EvoProDom presents a novel model for protein evolution based on the ‘mix and merge’ of protein domains rather than DNA-based evolution models. This confers the advantage of considering chromosomal alterations as drivers of protein evolutionary events.
中文翻译:

EvoProDom:通过评估蛋白质结构域的易位对蛋白质家族进行进化建模
在这里,我们介绍了一种新的“蛋白质结构域进化”(EvoProDom)模型,用于描述基于蛋白质结构域“混合和合并”的蛋白质进化。我们组装并整合了基因组和蛋白质组数据,包括来自 109 个生物体的蛋白质结构域内容和直系同源蛋白质。在 EvoProDom 中,我们将进化事件(特别是易位)描述为不同生物体中直系同源蛋白质之间蛋白质结构域的相互交换。我们表明,高频率易位的蛋白质结构域是由富含反式的转录物产生的。-剪接事件,即从两个不同基因的融合中产生新的转录本。在 EvoProDom 中,我们描述了一种使用 KoFamKOAL 和 Pfam 等工具整理来自 KEGG 的直系同源蛋白质注释和来自蛋白质序列的蛋白质结构域内容的通用方法。总而言之,EvoProDom 提出了一种新的蛋白质进化模型,它基于蛋白质结构域的“混合和合并”,而不是基于 DNA 的进化模型。这赋予了将染色体改变视为蛋白质进化事件驱动因素的优势。
更新日期:2021-09-01
中文翻译:

EvoProDom:通过评估蛋白质结构域的易位对蛋白质家族进行进化建模
在这里,我们介绍了一种新的“蛋白质结构域进化”(EvoProDom)模型,用于描述基于蛋白质结构域“混合和合并”的蛋白质进化。我们组装并整合了基因组和蛋白质组数据,包括来自 109 个生物体的蛋白质结构域内容和直系同源蛋白质。在 EvoProDom 中,我们将进化事件(特别是易位)描述为不同生物体中直系同源蛋白质之间蛋白质结构域的相互交换。我们表明,高频率易位的蛋白质结构域是由富含反式的转录物产生的。-剪接事件,即从两个不同基因的融合中产生新的转录本。在 EvoProDom 中,我们描述了一种使用 KoFamKOAL 和 Pfam 等工具整理来自 KEGG 的直系同源蛋白质注释和来自蛋白质序列的蛋白质结构域内容的通用方法。总而言之,EvoProDom 提出了一种新的蛋白质进化模型,它基于蛋白质结构域的“混合和合并”,而不是基于 DNA 的进化模型。这赋予了将染色体改变视为蛋白质进化事件驱动因素的优势。











































 京公网安备 11010802027423号
京公网安备 11010802027423号