Structure ( IF 4.4 ) Pub Date : 2021-06-30 , DOI: 10.1016/j.str.2021.06.007 Ashish A Kawale 1 , Björn M Burmann 1
|
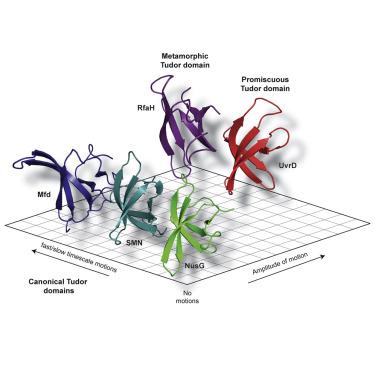
Tudor domains are crucial for mediating a diversity of protein-protein or protein-DNA interactions involved in nucleic acid metabolism. Using solution NMR spectroscopy, we assess the comprehensive understanding of the dynamical properties of the respective Tudor domains from four different bacterial (Escherichia coli) proteins UvrD, Mfd, RfaH, and NusG involved in different aspects of bacterial transcription regulation and associated processes. These proteins are benchmarked to the canonical Tudor domain fold from the human SMN protein. The detailed analysis of protein backbone dynamics and subsequent analysis by the Lipari-Szabo model-free approach revealed subtle differences in motions of the amide-bond vector on both pico- to nanosecond and micro- to millisecond timescales. On these timescales, our comparative approach reveals the usefulness of discrete amplitudes of dynamics to discern the different functionalities for Tudor domains exhibiting promiscuous binding, including the metamorphic Tudor domain included in the study.
中文翻译:

固有的骨架动力学微调都铎域的功能可塑性
Tudor 结构域对于介导参与核酸代谢的蛋白质-蛋白质或蛋白质-DNA 相互作用的多样性至关重要。使用溶液核磁共振光谱,我们评估了对四种不同细菌(大肠杆菌)各自 Tudor 结构域动力学特性的全面理解。) 蛋白质 UvrD、Mfd、RfaH 和 NusG 参与细菌转录调控和相关过程的不同方面。这些蛋白质以来自人类 SMN 蛋白质的典型 Tudor 结构域折叠为基准。蛋白质骨架动力学的详细分析和随后的 Lipari-Szabo 无模型方法分析揭示了酰胺键矢量在皮秒到纳秒和微秒到毫秒时间尺度上的运动的细微差异。在这些时间尺度上,我们的比较方法揭示了离散动力学幅度的有用性,以识别表现出混杂结合的 Tudor 域的不同功能,包括研究中包含的变质 Tudor 域。











































 京公网安备 11010802027423号
京公网安备 11010802027423号