当前位置:
X-MOL 学术
›
FEBS Open Bio
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A methylation-driven gene panel predicts survival in patients with colon cancer
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-06-29 , DOI: 10.1002/2211-5463.13242 Yaojun Peng 1, 2 , Jing Zhao 3 , Fan Yin 4 , Gaowa Sharen 5 , Qiyan Wu 6 , Qi Chen 7 , Xiaoxuan Sun 8, 9 , Juan Yang 10 , Huan Wang 3 , Dong Zhang 4
FEBS Open Bio ( IF 2.8 ) Pub Date : 2021-06-29 , DOI: 10.1002/2211-5463.13242 Yaojun Peng 1, 2 , Jing Zhao 3 , Fan Yin 4 , Gaowa Sharen 5 , Qiyan Wu 6 , Qi Chen 7 , Xiaoxuan Sun 8, 9 , Juan Yang 10 , Huan Wang 3 , Dong Zhang 4
Affiliation
|
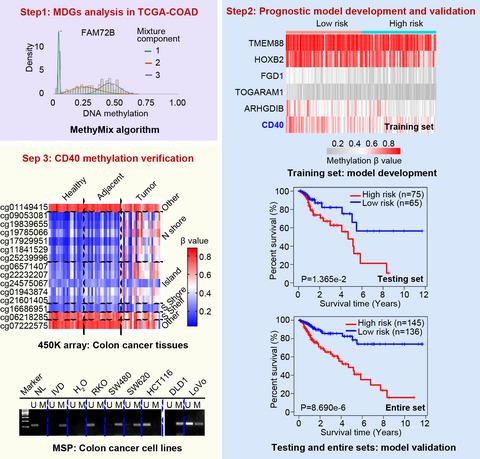
The accumulation of various genetic and epigenetic changes in colonic epithelial cells has been identified as one of the fundamental processes that drive the initiation and progression of colorectal cancer (CRC). This study aimed to explore functional genes regulated by DNA methylation and their potential utilization as biomarkers for the prediction of CRC prognoses. Methylation-driven genes (MDGs) were explored by applying the integrative analysis tool (methylmix) to The Cancer Genome Atlas CRC project. The prognostic MDG panel was identified by combining the Cox regression model with the least absolute shrinkage and selection operator regularization. Gene set enrichment analysis was used to determine the pathways associated with the six-MDG panel. Cluster of differentiation 40 (CD40) expression and methylation in CRC samples were validated by using additional datasets from the Gene Expression Omnibus. Methylation-specific PCR and bisulfite sequencing were used to confirm DNA methylation in CRC cell lines. A prognostic MDG panel consisting of six gene members was identified: TMEM88, HOXB2, FGD1, TOGARAM1, ARHGDIB and CD40. The high-risk phenotype classified by the six-MDG panel was associated with cancer-related biological processes, including invasion and metastasis, angiogenesis and the tumor immune microenvironment. The prognostic value of the six-MDG panel was found to be independent of tumor node metastasis stage and, in combination with tumor node metastasis stage and age, could help improve survival prediction. In addition, the expression of CD40 was confirmed to be regulated by promoter region methylation in CRC samples and cell lines. The proposed six-MDG panel represents a promising signature for estimating the prognosis of patients with CRC.
中文翻译:

甲基化驱动的基因组可预测结肠癌患者的生存率
结肠上皮细胞中各种遗传和表观遗传变化的积累已被确定为驱动结直肠癌 (CRC) 发生和进展的基本过程之一。本研究旨在探索受 DNA 甲基化调控的功能基因及其作为预测 CRC 预后的生物标志物的潜在用途。通过将综合分析工具 ( methylmix ) 应用于癌症基因组图谱 CRC 项目,探索了甲基化驱动基因 (MDG) 。通过将 Cox 回归模型与最小绝对收缩和选择算子正则化相结合来确定预后 MDG 面板。基因集富集分析用于确定与六千年发展目标相关的途径。分化簇 40 ( CD40) 通过使用来自 Gene Expression Omnibus 的其他数据集验证 CRC 样本中的表达和甲基化。甲基化特异性 PCR 和亚硫酸氢盐测序用于确认 CRC 细胞系中的 DNA 甲基化。确定了由六个基因成员组成的预后 MDG 面板:TMEM88、HOXB2、FGD1、TOGARAM1、ARHGDIB和CD40. 6-MDG 小组分类的高危表型与癌症相关的生物学过程相关,包括侵袭和转移、血管生成和肿瘤免疫微环境。发现 6-MDG 小组的预后价值与肿瘤淋巴结转移阶段无关,结合肿瘤淋巴结转移阶段和年龄,有助于提高生存预测。此外,在 CRC 样品和细胞系中,CD40的表达被证实受启动子区域甲基化的调节。拟议的六千年发展目标小组代表了估计CRC患者预后的有希望的标志。
更新日期:2021-09-01
中文翻译:

甲基化驱动的基因组可预测结肠癌患者的生存率
结肠上皮细胞中各种遗传和表观遗传变化的积累已被确定为驱动结直肠癌 (CRC) 发生和进展的基本过程之一。本研究旨在探索受 DNA 甲基化调控的功能基因及其作为预测 CRC 预后的生物标志物的潜在用途。通过将综合分析工具 ( methylmix ) 应用于癌症基因组图谱 CRC 项目,探索了甲基化驱动基因 (MDG) 。通过将 Cox 回归模型与最小绝对收缩和选择算子正则化相结合来确定预后 MDG 面板。基因集富集分析用于确定与六千年发展目标相关的途径。分化簇 40 ( CD40) 通过使用来自 Gene Expression Omnibus 的其他数据集验证 CRC 样本中的表达和甲基化。甲基化特异性 PCR 和亚硫酸氢盐测序用于确认 CRC 细胞系中的 DNA 甲基化。确定了由六个基因成员组成的预后 MDG 面板:TMEM88、HOXB2、FGD1、TOGARAM1、ARHGDIB和CD40. 6-MDG 小组分类的高危表型与癌症相关的生物学过程相关,包括侵袭和转移、血管生成和肿瘤免疫微环境。发现 6-MDG 小组的预后价值与肿瘤淋巴结转移阶段无关,结合肿瘤淋巴结转移阶段和年龄,有助于提高生存预测。此外,在 CRC 样品和细胞系中,CD40的表达被证实受启动子区域甲基化的调节。拟议的六千年发展目标小组代表了估计CRC患者预后的有希望的标志。











































 京公网安备 11010802027423号
京公网安备 11010802027423号