Cell Chemical Biology ( IF 6.6 ) Pub Date : 2021-06-28 , DOI: 10.1016/j.chembiol.2021.05.022 Alison C Waldman 1 , Balaji M Rao 2 , Albert J Keung 1
|
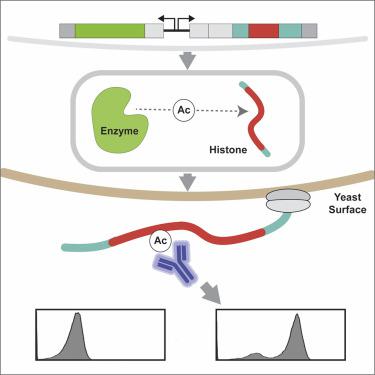
Histone proteins are decorated with a combinatorially and numerically diverse set of biochemical modifications. Here, we describe a versatile and scalable approach which enables efficient characterization of histone modifications without the need for recombinant protein production. As proof-of-concept, we first use this system to rapidly profile the histone H3 and H4 residue writing specificities of the human histone acetyltransferase, p300. Subsequently, a large panel of commercially available anti-acetylation antibodies are screened for their specificities, identifying many suitable and unsuitable reagents. Furthermore, this approach enables efficient mapping of the large binary crosstalk space between acetylated residues on histones H3 and H4 and uncovers residue interdependencies affecting p300 activity. These results show that using yeast surface display to study histone modifications is a useful tool that can advance our understanding of chromatin biology by enabling efficient interrogation of the complexity of epigenome modifications.
中文翻译:

通过酵母表面展示绘制表观基因组酶的残基特异性
组蛋白装饰有一组组合和数量不同的生化修饰。在这里,我们描述了一种通用且可扩展的方法,该方法无需重组蛋白生产即可有效表征组蛋白修饰。作为概念验证,我们首先使用该系统快速分析人类组蛋白乙酰转移酶 p300 的组蛋白 H3 和 H4 残基写入特异性。随后,对大量市售抗乙酰化抗体的特异性进行筛选,确定许多合适和不合适的试剂。此外,这种方法能够有效映射组蛋白 H3 和 H4 上乙酰化残基之间的大二元串扰空间,并揭示影响 p300 活性的残基相互依赖性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号