当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Gene Expression Biomarker Predicts Heat Shock Factor 1 Activation in a Gene Expression Compendium
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2021-06-25 , DOI: 10.1021/acs.chemrestox.0c00510 Patrick W Cervantes 1, 2 , J Christopher Corton 1
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2021-06-25 , DOI: 10.1021/acs.chemrestox.0c00510 Patrick W Cervantes 1, 2 , J Christopher Corton 1
Affiliation
|
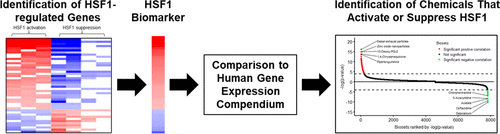
The United States Environmental Protection Agency (US EPA) recently developed a tiered testing strategy to use advances in high-throughput transcriptomics (HTTr) testing to identify molecular targets of thousands of environmental chemicals that can be linked to adverse outcomes. Here, we describe a method that uses a gene expression biomarker to predict chemical activation of heat shock factor 1 (HSF1), a transcription factor critical for proteome maintenance. The HSF1 biomarker was built from transcript profiles derived from A375 cells exposed to a HSF1-activating heat shock protein (HSP) 90 inhibitor in the presence or absence of HSF1 expression. The resultant 44 identified genes included those that (1) are dependent on HSF1 for regulation, (2) have direct interactions with HSF1 assessed by ChIP-Seq, and (3) are in the molecular chaperone family. To test for accuracy, the biomarker was compared in a pairwise manner to gene lists derived from treatments with known HSF1 activity (HSP and proteasomal inhibitors) using the correlation-based Running Fisher test; the balanced accuracy for prediction was 96%. A microarray compendium consisting of 12,092 microarray comparisons from human cells exposed to 2670 individual chemicals was screened using our approach; 112 and 19 chemicals were identified as putative HSF1 activators or suppressors, respectively, and most appear to be novel modulators. A large percentage of the chemical treatments that induced HSF1 also induced oxidant-activated NRF2 (∼46%). For five compounds or mixtures, we found that NRF2 activation occurred at lower concentrations or at earlier times than HSF1 activation, supporting the concept of a tiered cellular protection system dependent on the level of chemical-induced stress. The approach described here could be used to identify environmentally relevant chemical HSF1 activators in HTTr data sets.
中文翻译:

基因表达生物标志物可预测基因表达纲要中的热休克因子 1 激活
美国环境保护署 (US EPA) 最近制定了一种分层测试策略,以利用高通量转录组学 (HTTr) 测试的进步来识别可能与不良结果相关的数千种环境化学物质的分子靶标。在这里,我们描述了一种使用基因表达生物标志物来预测热休克因子 1 (HSF1) 化学激活的方法,热休克因子 1 (HSF1) 是一种对蛋白质组维持至关重要的转录因子。HSF1 生物标志物是根据 A375 细胞的转录谱构建的,这些细胞在 HSF1 表达存在或不存在的情况下暴露于 HSF1 激活热休克蛋白 (HSP) 90 抑制剂。由此产生的 44 个已识别基因包括 (1) 依赖 HSF1 进行调节,(2) 与 ChIP-Seq 评估的 HSF1 直接相互作用,以及 (3) 属于分子伴侣家族。为了测试准确性,使用基于相关性的 Running Fisher 测试,以成对方式将生物标志物与源自具有已知 HSF1 活性(HSP 和蛋白酶体抑制剂)的治疗的基因列表进行比较;预测的平衡准确率为 96%。使用我们的方法筛选了由暴露于 2670 种化学物质的人类细胞的 12,092 个微阵列比较组成的微阵列纲要;112 和 19 种化学物质分别被确定为推定的 HSF1 激活剂或抑制剂,并且大多数似乎是新型调节剂。大部分诱导 HSF1 的化学处理也诱导了氧化剂激活的 NRF2(~46%)。对于五种化合物或混合物,我们发现 NRF2 激活发生在比 HSF1 激活更低的浓度或更早的时间,支持依赖于化学诱导压力水平的分层细胞保护系统的概念。此处描述的方法可用于识别 HTTr 数据集中与环境相关的化学 HSF1 激活剂。
更新日期:2021-07-19
中文翻译:

基因表达生物标志物可预测基因表达纲要中的热休克因子 1 激活
美国环境保护署 (US EPA) 最近制定了一种分层测试策略,以利用高通量转录组学 (HTTr) 测试的进步来识别可能与不良结果相关的数千种环境化学物质的分子靶标。在这里,我们描述了一种使用基因表达生物标志物来预测热休克因子 1 (HSF1) 化学激活的方法,热休克因子 1 (HSF1) 是一种对蛋白质组维持至关重要的转录因子。HSF1 生物标志物是根据 A375 细胞的转录谱构建的,这些细胞在 HSF1 表达存在或不存在的情况下暴露于 HSF1 激活热休克蛋白 (HSP) 90 抑制剂。由此产生的 44 个已识别基因包括 (1) 依赖 HSF1 进行调节,(2) 与 ChIP-Seq 评估的 HSF1 直接相互作用,以及 (3) 属于分子伴侣家族。为了测试准确性,使用基于相关性的 Running Fisher 测试,以成对方式将生物标志物与源自具有已知 HSF1 活性(HSP 和蛋白酶体抑制剂)的治疗的基因列表进行比较;预测的平衡准确率为 96%。使用我们的方法筛选了由暴露于 2670 种化学物质的人类细胞的 12,092 个微阵列比较组成的微阵列纲要;112 和 19 种化学物质分别被确定为推定的 HSF1 激活剂或抑制剂,并且大多数似乎是新型调节剂。大部分诱导 HSF1 的化学处理也诱导了氧化剂激活的 NRF2(~46%)。对于五种化合物或混合物,我们发现 NRF2 激活发生在比 HSF1 激活更低的浓度或更早的时间,支持依赖于化学诱导压力水平的分层细胞保护系统的概念。此处描述的方法可用于识别 HTTr 数据集中与环境相关的化学 HSF1 激活剂。











































 京公网安备 11010802027423号
京公网安备 11010802027423号