当前位置:
X-MOL 学术
›
J. Appl. Ecol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Building an ecologically founded disease risk prioritization framework for migratory wildlife species based on contact with livestock
Journal of Applied Ecology ( IF 5.0 ) Pub Date : 2021-05-31 , DOI: 10.1111/1365-2664.13937 Munib Khanyari 1, 2, 3 , Sarah Robinson 2 , Eric R. Morgan 1, 4 , Tony Brown 4 , Navinder J. Singh 5 , Albert Salemgareyev 6 , Steffen Zuther 6, 7 , Richard Kock 8 , E. J. Milner‐Gulland 2
中文翻译:

基于与牲畜接触的迁徙野生动物物种建立以生态为基础的疾病风险优先级框架
更新日期:2021-05-31
Journal of Applied Ecology ( IF 5.0 ) Pub Date : 2021-05-31 , DOI: 10.1111/1365-2664.13937 Munib Khanyari 1, 2, 3 , Sarah Robinson 2 , Eric R. Morgan 1, 4 , Tony Brown 4 , Navinder J. Singh 5 , Albert Salemgareyev 6 , Steffen Zuther 6, 7 , Richard Kock 8 , E. J. Milner‐Gulland 2
Affiliation
|
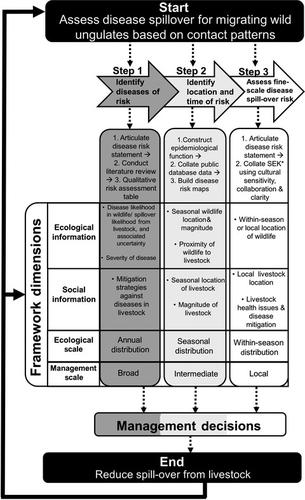
- Shared use of rangelands by livestock and wildlife can lead to disease transmission. To align agricultural livelihoods with wildlife conservation, a multipronged and interdisciplinary approach for disease management is needed, particularly in data-limited situations with migratory hosts. Migratory wildlife and livestock can range over vast areas, and opportunities for disease control interventions are limited. Predictive frameworks are needed which can allow for identification of potential sites and timings of interventions.
- We developed an iterative three-step framework to assess cross-species disease transmission risk between migrating wildlife and livestock in data-limited circumstances and across social-ecological scales. The framework first assesses risk of transmission for potentially important diseases for hosts in a multi-use landscape. Following this, it uses an epidemiological risk function to represent transmission-relevant contact patterns, using density and distribution of the host to map locations and periods of disease risk. Finally, it takes fine-scale data on livestock management and observed wildlife–livestock interactions to provide locally relevant insights on disease risk.
- We applied the framework to characterize disease transmission between livestock and saiga antelopes Saiga tatarica in Central Kazakhstan.
- At step 1, we identified peste-des-petits-ruminants as posing a high risk of transmission from livestock to saigas, foot-and-mouth disease as low risk, lumpy skin disease as unknown and pasteurellosis as uncertain risk. At step 2, we identified regions of high disease transmission risk at different times of year, indicating where disease management should be focussed. At step 3, we synthesized field surveys, government data and literature review to assess the role of livestock in the 2015 saiga mass mortality event from pasteurellosis, concluding that it was minimal.
- Synthesis and applications. Our iterative framework has wide applicability in assessing and predicting disease spill-over at management-relevant temporal and spatial scales in areas where livestock share space with migratory species. Our case study demonstrated the value of combining ecological and social information to inform management of targeted interventions to reduce disease risk, which can be used to plan disease surveillance and vaccination programmes.
中文翻译:

基于与牲畜接触的迁徙野生动物物种建立以生态为基础的疾病风险优先级框架
- 牲畜和野生动物共用牧场会导致疾病传播。为了使农业生计与野生动物保护保持一致,需要多管齐下和跨学科的疾病管理方法,尤其是在迁徙宿主数据有限的情况下。迁徙的野生动物和牲畜可以分布在广阔的地区,疾病控制干预的机会有限。需要能够识别潜在干预地点和时间的预测框架。
- 我们开发了一个迭代的三步框架,以评估在数据有限的情况下以及跨社会生态尺度迁移的野生动物和牲畜之间的跨物种疾病传播风险。该框架首先评估了多用途景观中宿主潜在重要疾病的传播风险。在此之后,它使用流行病学风险函数来表示与传播相关的接触模式,使用宿主的密度和分布来绘制疾病风险的位置和时期。最后,它需要牲畜管理的精细数据和观察到的野生动物-牲畜相互作用,以提供有关疾病风险的当地相关见解。
- 我们应用该框架来描述哈萨克斯坦中部的牲畜和赛加羚羊Saiga tatarica之间的疾病传播特征。
- 在第 1 步,我们确定了小反刍动物疫病从牲畜传播到赛加羚羊的高风险、低风险的口蹄疫、未知的肿块皮肤病和不确定的巴氏杆菌病。在第 2 步中,我们确定了一年中不同时间的疾病传播高风险区域,指明了疾病管理的重点。在第 3 步,我们综合了实地调查、政府数据和文献综述,以评估牲畜在 2015 年巴氏杆菌病导致的赛加羚羊大规模死亡事件中的作用,得出的结论是这种影响很小。
- 合成与应用。在牲畜与迁徙物种共享空间的地区,我们的迭代框架在评估和预测与管理相关的时间和空间尺度上的疾病溢出方面具有广泛的适用性。我们的案例研究证明了将生态和社会信息结合起来为管理有针对性的干预措施以降低疾病风险提供信息的价值,这些干预措施可用于规划疾病监测和疫苗接种计划。











































 京公网安备 11010802027423号
京公网安备 11010802027423号