当前位置:
X-MOL 学术
›
Microbiologyopen
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Optimized bacterial DNA isolation method for microbiome analysis of human tissues
MicrobiologyOpen ( IF 3.9 ) Pub Date : 2021-06-16 , DOI: 10.1002/mbo3.1191 Carlijn E Bruggeling 1 , Daniel R Garza 2, 3 , Soumia Achouiti 1 , Wouter Mes 4, 5 , Bas E Dutilh 2, 6 , Annemarie Boleij 1
MicrobiologyOpen ( IF 3.9 ) Pub Date : 2021-06-16 , DOI: 10.1002/mbo3.1191 Carlijn E Bruggeling 1 , Daniel R Garza 2, 3 , Soumia Achouiti 1 , Wouter Mes 4, 5 , Bas E Dutilh 2, 6 , Annemarie Boleij 1
Affiliation
|
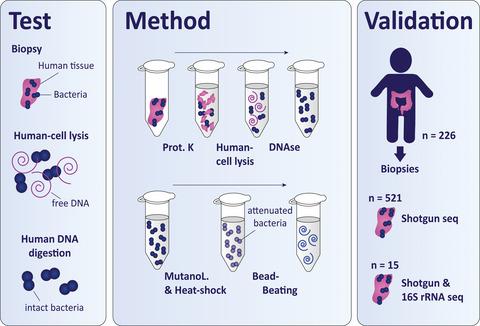
Recent advances in microbiome sequencing have rendered new insights into the role of the microbiome in human health with potential clinical implications. Unfortunately, the presence of host DNA in tissue isolates has hampered the analysis of host-associated bacteria. Here, we present a DNA isolation protocol for tissue, optimized on biopsies from resected human colons (~2–5 mm in size), which includes reduction of human DNA without distortion of relative bacterial abundance at the phylum level. We evaluated which concentrations of Triton and saponin lyse human cells and leave bacterial cells intact, in combination with DNAse treatment to deplete released human DNA. Saponin at a concentration of 0.0125% in PBS lysed host cells, resulting in a 4.5-fold enrichment of bacterial DNA while preserving the relative abundance of Firmicutes, Bacteroidetes, γ-Proteobacteria, and Actinobacteria assessed by qPCR. Our optimized protocol was validated in the setting of two large clinical studies on 521 in vivo acquired colon biopsies of 226 patients using shotgun metagenomics. The resulting bacterial profiles exhibited alpha and beta diversities that are similar to the diversities found by 16S rRNA amplicon sequencing. A direct comparison between shotgun metagenomics and 16S rRNA amplicon sequencing of 15 forceps tissue biopsies showed similar bacterial profiles and a similar Shannon diversity index between the sequencing methods. Hereby, we present the first protocol for enriching bacterial DNA from tissue biopsies that allows efficient isolation of all bacteria. Our protocol facilitates analysis of a wide spectrum of bacteria of clinical tissue samples improving their applicability for microbiome research.
中文翻译:

用于人体组织微生物组分析的优化细菌 DNA 分离方法
微生物组测序的最新进展为微生物组在人类健康中的作用提供了新的见解,并具有潜在的临床意义。不幸的是,组织分离物中宿主 DNA 的存在阻碍了对宿主相关细菌的分析。在这里,我们提出了一种组织 DNA 分离方案,该方案针对切除的人类结肠(约 2-5 毫米大小)的活检进行了优化,其中包括减少人类 DNA,而不会在门水平上扭曲相对细菌丰度。我们评估了哪些浓度的 Triton 和皂苷溶解人体细胞并保持细菌细胞完整,并结合 DNAse 处理以消耗释放的人体 DNA。在 PBS 裂解的宿主细胞中浓度为 0.0125% 的皂苷,导致细菌 DNA 富集 4.5 倍,同时保持相对丰度厚壁菌门、拟杆菌门、γ-变形菌门和放线菌门通过 qPCR 评估。我们的优化方案在两项大型临床研究的设置中得到验证,该研究使用鸟枪法宏基因组学对 226 名患者的 521 例体内获得性结肠活检进行了验证。由此产生的细菌谱显示出与 16S rRNA 扩增子测序发现的多样性相似的 alpha 和 beta 多样性。鸟枪法宏基因组学和 15 个镊子组织活检的 16S rRNA 扩增子测序之间的直接比较显示,测序方法之间的细菌谱和香农多样性指数相似。因此,我们提出了第一个从组织活检中富集细菌 DNA 的协议,该协议允许有效分离所有细菌。我们的协议有助于分析临床组织样本中的多种细菌,从而提高它们对微生物组研究的适用性。
更新日期:2021-06-17
中文翻译:

用于人体组织微生物组分析的优化细菌 DNA 分离方法
微生物组测序的最新进展为微生物组在人类健康中的作用提供了新的见解,并具有潜在的临床意义。不幸的是,组织分离物中宿主 DNA 的存在阻碍了对宿主相关细菌的分析。在这里,我们提出了一种组织 DNA 分离方案,该方案针对切除的人类结肠(约 2-5 毫米大小)的活检进行了优化,其中包括减少人类 DNA,而不会在门水平上扭曲相对细菌丰度。我们评估了哪些浓度的 Triton 和皂苷溶解人体细胞并保持细菌细胞完整,并结合 DNAse 处理以消耗释放的人体 DNA。在 PBS 裂解的宿主细胞中浓度为 0.0125% 的皂苷,导致细菌 DNA 富集 4.5 倍,同时保持相对丰度厚壁菌门、拟杆菌门、γ-变形菌门和放线菌门通过 qPCR 评估。我们的优化方案在两项大型临床研究的设置中得到验证,该研究使用鸟枪法宏基因组学对 226 名患者的 521 例体内获得性结肠活检进行了验证。由此产生的细菌谱显示出与 16S rRNA 扩增子测序发现的多样性相似的 alpha 和 beta 多样性。鸟枪法宏基因组学和 15 个镊子组织活检的 16S rRNA 扩增子测序之间的直接比较显示,测序方法之间的细菌谱和香农多样性指数相似。因此,我们提出了第一个从组织活检中富集细菌 DNA 的协议,该协议允许有效分离所有细菌。我们的协议有助于分析临床组织样本中的多种细菌,从而提高它们对微生物组研究的适用性。










































 京公网安备 11010802027423号
京公网安备 11010802027423号