当前位置:
X-MOL 学术
›
Chem. Bio. Drug Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
On the use of electronegativity and electron affinity based pseudo-molecular field descriptors in developing correlations for quantitative structure-activity relationship modeling of drug activities
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2021-05-19 , DOI: 10.1111/cbdd.13895 Pushkar D Kunde 1, 2 , Sudha Ramkumar 3 , Sanjay P Kamble 1, 2 , Ameeta Ravikumar 4 , Bhaskar D Kulkarni 1, 2 , V Ravi Kumar 1, 2
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2021-05-19 , DOI: 10.1111/cbdd.13895 Pushkar D Kunde 1, 2 , Sudha Ramkumar 3 , Sanjay P Kamble 1, 2 , Ameeta Ravikumar 4 , Bhaskar D Kulkarni 1, 2 , V Ravi Kumar 1, 2
Affiliation
|
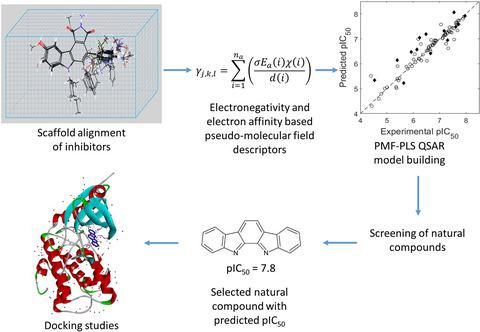
For quantitative structure-activity relationship (QSAR) modeling in ligand-based drug discovery programs, pseudo-molecular field (PMF) descriptors using intrinsic atomic properties, namely, electronegativity and electron affinity are studied. In combination with partial least squares analysis and Procrustes transformation, these PMF descriptors were employed successfully to develop correlations that predict the activities of target protein inhibitors involved in various diseases (cancer, neurodegenerative disorders, HIV, and malaria). The results show that the present QSAR approach is competitive to existing QSAR models. In order to demonstrate the use of this algorithm, we present results of screening naturally occurring molecules with unknown bioactivities. The pIC50 predictions can screen molecules that have desirable activity before assessment by docking studies.
中文翻译:

基于电负性和电子亲和性的伪分子场描述符在开发药物活性定量构效关系模型的相关性中的应用
对于基于配体的药物发现程序中的定量构效关系 (QSAR) 建模,研究了使用固有原子特性(即电负性和电子亲和力)的伪分子场 (PMF) 描述符。结合偏最小二乘分析和 Procrustes 变换,这些 PMF 描述符被成功用于开发相关性,以预测与各种疾病(癌症、神经退行性疾病、HIV 和疟疾)相关的靶蛋白抑制剂的活性。结果表明,目前的 QSAR 方法与现有的 QSAR 模型相比具有竞争力。为了演示该算法的使用,我们展示了筛选具有未知生物活性的天然存在的分子的结果。PIC 50 在通过对接研究进行评估之前,预测可以筛选具有所需活性的分子。
更新日期:2021-07-14
中文翻译:

基于电负性和电子亲和性的伪分子场描述符在开发药物活性定量构效关系模型的相关性中的应用
对于基于配体的药物发现程序中的定量构效关系 (QSAR) 建模,研究了使用固有原子特性(即电负性和电子亲和力)的伪分子场 (PMF) 描述符。结合偏最小二乘分析和 Procrustes 变换,这些 PMF 描述符被成功用于开发相关性,以预测与各种疾病(癌症、神经退行性疾病、HIV 和疟疾)相关的靶蛋白抑制剂的活性。结果表明,目前的 QSAR 方法与现有的 QSAR 模型相比具有竞争力。为了演示该算法的使用,我们展示了筛选具有未知生物活性的天然存在的分子的结果。PIC 50 在通过对接研究进行评估之前,预测可以筛选具有所需活性的分子。











































 京公网安备 11010802027423号
京公网安备 11010802027423号