Current Bioinformatics ( IF 2.4 ) Pub Date : 2021-02-28 , DOI: 10.2174/1574893615999200715165335 Qingqi Zhu 1 , Yongxian Fan 1 , Xiaoyong Pan 2
|
Background: MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs with about 22 nucleotides, and they play a significant role in a variety of complex biological processes. Many researches have shown that miRNAs are closely related to human diseases. Although the biological experiments are reliable in identifying miRNA-disease associations, they are timeconsuming and costly.
Objective: Thus, computational methods are urgently needed to effectively predict miRNA-disease associations.
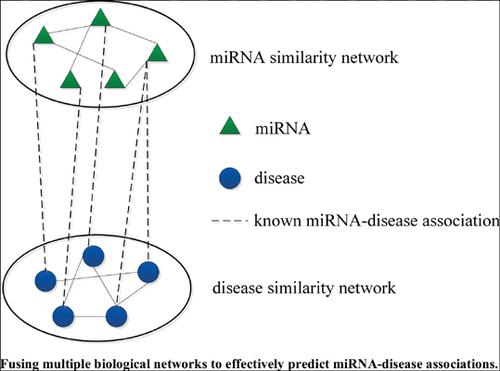
Methods: In this paper, we proposed a novel method, BIRWMDA, based on a bi-random walk model to predict miRNA-disease associations. Specifically, in BIRWMDA, the similarity network fusion algorithm is used to combine the multiple similarity matrices to obtain a miRNA-miRNA similarity matrix and a disease-disease similarity matrix, then the miRNA-disease associations were predicted by the bi-random walk model.
Results: To evaluate the performance of BIRWMDA, we ran the leave-one-out cross-validation and 5-fold cross-validation, and their corresponding AUCs were 0.9303 and 0.9223 ± 0.00067, respectively. To further demonstrate the effectiveness of the BIRWMDA, from the perspective of exploring disease-related miRNAs, we conducted three case studies of breast neoplasms, prostate neoplasms and gastric neoplasms, where 48, 50 and 50 out of the top 50 predicted miRNAs were confirmed by literature, respectively. From the perspective of exploring miRNA-related diseases, we conducted two case studies of hsa-mir-21 and hsa-mir-155, where 7 and 5 out of the top 10 predicted diseases were confirmed by literatures, respectively.
Conclusion: The fusion of multiple biological networks could effectively predict miRNA-diseases associations. We expected BIRWMDA to serve as a biological tool for mining potential miRNAdisease associations.
中文翻译:

融合多个生物网络以有效预测miRNA疾病关联。
背景:MicroRNA(miRNA)是一类具有约22个核苷酸的内源性非编码RNA,它们在各种复杂的生物过程中均起着重要作用。许多研究表明,miRNA与人类疾病密切相关。尽管生物学实验在鉴定miRNA-疾病关联方面是可靠的,但它们既耗时又昂贵。
目的:因此,迫切需要一种计算方法来有效预测miRNA-疾病关联。
方法:在本文中,我们提出了一种基于双向步行模型的新方法BIRWMDA,用于预测miRNA-疾病关联。具体而言,在BIRWMDA中,使用相似性网络融合算法将多个相似性矩阵组合以获得miRNA-miRNA相似性矩阵和疾病-疾病相似性矩阵,然后通过双随机行走模型预测miRNA-疾病关联。
结果:为了评估BIRWMDA的性能,我们进行了留一法交叉验证和5倍交叉验证,其相应的AUC分别为0.9303和0.9223±0.00067。为了进一步证明BIRWMDA的有效性,从探索与疾病相关的miRNA的角度出发,我们对乳腺肿瘤,前列腺肿瘤和胃肿瘤进行了三个案例研究,其中前50个预测的miRNA中有48个,50个和50个被证实。文学。从探索与miRNA相关的疾病的角度出发,我们进行了hsa-mir-21和hsa-mir-155的两个案例研究,在前十种预测疾病中分别有7例和5例得到了文献证实。
结论:多种生物网络的融合可以有效地预测miRNA疾病的关联。我们期望BIRWMDA可以作为挖掘潜在的miRNA疾病关联的生物学工具。










































 京公网安备 11010802027423号
京公网安备 11010802027423号