Current Bioinformatics ( IF 2.4 ) Pub Date : 2021-02-28 , DOI: 10.2174/1574893615999200724145434 Nikhila T Suresh 1 , Vimina E R 1 , U. Krishnakumar 1
|
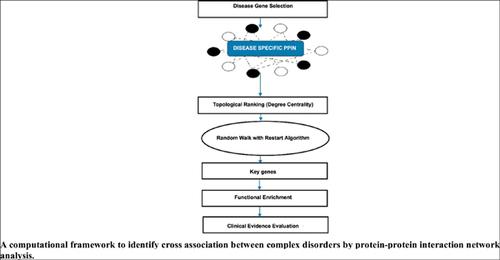
Objective: It is a known fact that numerous complex disorders do not happen in isolation indicating the plausible set of shared causes common to several different sicknesses. Hence, analysis of comorbidity can be utilized to explore the association between several disorders. In this study, we have proposed a network-based computational approach, in which genes are organized based on the topological characteristics of the constructed Protein-Protein Interaction Network (PPIN) followed by a network prioritization scheme, to identify distinctive key genes and biological pathways shared among diseases.
Methods: The proposed approach is initiated from constructed PPIN of any randomly chosen disease genes in order to infer its associations with other diseases in terms of shared pathways, coexpression, co-occurrence etc. For this, initially, proteins associated to any disease based on random choice were identified. Secondly, PPIN is organized through topological analysis to define hub genes. Finally, using a prioritization algorithm a ranked list of newly predicted multimorbidity-associated proteins is generated. Using Gene Ontology (GO), cellular pathways involved in multimorbidity-associated proteins are mined.
Result and Conclusion: The proposed methodology is tested using three disorders, namely Diabetes, Obesity and blood pressure at an atomic level and the results suggest the comorbidity of other complex diseases that have associations with the proteins included in the disease of present study through shared proteins and pathways. For diabetes, we have obtained key genes like GAPDH, TNF, IL6, AKT1, ALB, TP53, IL10, MAPK3, TLR4 and EGF with key pathways like P53 pathway, VEGF signaling pathway, Ras Pathway, Interleukin signaling pathway, Endothelin signaling pathway, Huntington disease etc. Studies on other disorders such as obesity and blood pressure also revealed promising results.
中文翻译:

通过蛋白质-蛋白质相互作用网络分析识别复杂疾病之间的交叉关联的计算框架
目的:众所周知的事实是,许多复杂的疾病并未单独发生,这表明几种不同疾病共有的共同原因似乎是合理的。因此,合并症的分析可以用来探索几种疾病之间的关联。在这项研究中,我们提出了一种基于网络的计算方法,其中,根据所构建的蛋白质-蛋白质相互作用网络(PPIN)的拓扑特征组织基因,然后根据网络优先顺序排列方案,以识别独特的关键基因和生物学途径在疾病之间共享。
方法:所提出的方法是从构建的任意随机选择的疾病基因的PPIN起始的,以便从共享途径,共表达,共现等方面推断其与其他疾病的关联。为此,最初,与任何疾病相关的蛋白质均基于确定了随机选择。其次,通过拓扑分析来组织PPIN,以定义集线器基因。最后,使用优先级排序算法,将生成新预测的多发病相关蛋白的排名列表。使用基因本体论(GO),可以发现与多发病相关蛋白有关的细胞途径。
结果与结论:所提出的方法在三种疾病上进行了测试,即糖尿病,肥胖和血压在原子水平上,结果表明其他复杂疾病的合并症与通过共享蛋白质与本研究疾病所含蛋白质相关联和途径。对于糖尿病,我们获得了关键基因,如GAPDH,TNF,IL6,AKT1,ALB,TP53,IL10,MAPK3,TLR4和EGF,其关键途径包括P53途径,VEGF信号传导途径,Ras途径,白介素信号传导途径,内皮素信号传导途径,亨廷顿病等。对肥胖和血压等其他疾病的研究也显示出令人鼓舞的结果。











































 京公网安备 11010802027423号
京公网安备 11010802027423号