Current Bioinformatics ( IF 4 ) Pub Date : 2021-02-28 , DOI: 10.2174/1574893615999200707142852 Syed Adnan Shah Bukhari 1 , Abdul Razzaq 1 , Javeria Jabeen 1 , Shaheer Khan 1 , Zulqurnain Khan 2
|
Background: With the rapid development of the sequencing methods in recent years, binding sites have been systematically identified in such projects as Nested-MICA and MEME. Prediction of DNA motifs with higher accuracy and precision has been a very important task for bioinformaticians. Nevertheless, experimental approaches are still time-consuming for big data set, making computational identification of binding sites indispensable.
Objective: To facilitate the identification of the binding site, we proposed a deep learning architecture, named Deep-BSC (Deep-Learning Binary Search Classification), to predict binding sites in a raw DNA sequence with more precision and accuracy.
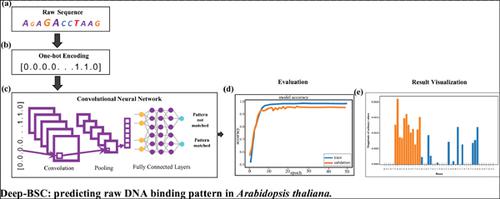
Methods: Our proposed architecture purely relies on the raw DNA sequence to predict the binding sites for protein by using a convolutional neural network (CNN). We trained our deep learning model on binding sites at the nucleotide level. DNA sequence of A. thaliana is used in this study because it is a model plant.
Results: The results demonstrate the effectiveness and efficiency of our method in the classification of binding sites against random sequences, using deep learning. We construct a CNN with different layers and filters to show the usefulness of max-pooling technique in the proposed method. To gain the interpretability of our approach, we further visualized binding sites in the saliency map and successfully identified similar motifs in the raw sequence. The proposed computational framework is time and resource efficient.
Conclusion: Deep-BSC enables the identification of binding sites in the DNA sequences via a highly accurate CNN. The proposed computational framework can also be applied to problems such as operator, repeats in the genome, DNA markers, and recognition sites for enzymes, thereby promoting the use of Deep-BSC method in life sciences.
中文翻译:

Deep-BSC:预测拟南芥中原始的DNA结合模式
背景:随着近年来测序方法的飞速发展,在Nested-MICA和MEME等项目中系统地确定了结合位点。对于生物信息学家来说,以更高的准确度和精确度预测DNA基序一直是非常重要的任务。然而,对于大数据集,实验方法仍然很耗时,从而使结合位点的计算鉴定成为必不可少的。
目的:为促进结合位点的鉴定,我们提出了一种名为Deep-BSC(深度学习二进制搜索分类)的深度学习架构,可以更精确,更准确地预测原始DNA序列中的结合位点。
方法:我们提出的体系结构完全依靠原始DNA序列通过使用卷积神经网络(CNN)来预测蛋白质的结合位点。我们在核苷酸水平的结合位点上训练了我们的深度学习模型。本研究使用拟南芥的DNA序列,因为它是模型植物。
结果:结果表明,使用深度学习,我们的方法在针对随机序列的结合位点分类中的有效性和效率。我们构造了具有不同层和滤波器的CNN,以显示所提出的方法中最大池技术的有用性。为了获得我们方法的可解释性,我们进一步在显着图中可视化了结合位点,并成功地在原始序列中鉴定出相似的基序。所提出的计算框架节省了时间和资源。
结论:Deep-BSC可以通过高度精确的CNN识别DNA序列中的结合位点。提出的计算框架还可以应用于诸如操作员,基因组重复,DNA标记和酶识别位点之类的问题,从而促进Deep-BSC方法在生命科学中的使用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号