当前位置:
X-MOL 学术
›
Genes Cells
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Transcriptome analysis of gene expression changes upon enzymatic dissociation in skeletal myoblasts
Genes to Cells ( IF 1.3 ) Pub Date : 2021-05-13 , DOI: 10.1111/gtc.12870 Atsuko Miyawaki-Kuwakado 1 , Qianmei Wu 1 , Akihito Harada 1 , Kosuke Tomimatsu 1 , Takeru Fujii 1 , Kazumitsu Maehara 1 , Yasuyuki Ohkawa 1
Genes to Cells ( IF 1.3 ) Pub Date : 2021-05-13 , DOI: 10.1111/gtc.12870 Atsuko Miyawaki-Kuwakado 1 , Qianmei Wu 1 , Akihito Harada 1 , Kosuke Tomimatsu 1 , Takeru Fujii 1 , Kazumitsu Maehara 1 , Yasuyuki Ohkawa 1
Affiliation
|
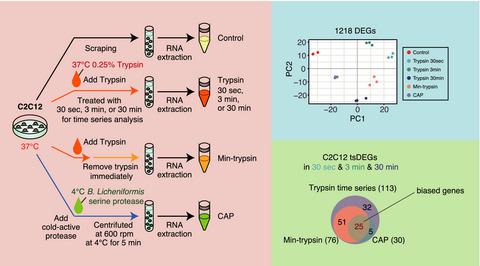
Single-cell RNA-sequencing analysis is one of the most effective tools for understanding specific cellular states. The use of single cells or pooled cells in RNA-seq analysis requires the isolation of cells from a tissue or culture. Although trypsin or more recently cold-active protease (CAP) has been used for cell dissociation, the extent to which the gene expression changes are suppressed has not been clarified. To this end, we conducted detailed profiling of the enzyme-dependent gene expression changes in mouse skeletal muscle progenitor cells, focusing on the enzyme treatment time, amount and temperature. We found that the genes whose expression was changed by the enzyme treatment could be classified in a time-dependent manner and that there were genes whose expression was changed independently of the enzyme treatment time, amount and temperature. This study will be useful as reference data for genes that should be excluded or considered for RNA-seq analysis using enzyme isolation methods.
中文翻译:

骨骼成肌细胞酶解后基因表达变化的转录组分析
单细胞 RNA 测序分析是了解特定细胞状态的最有效工具之一。在 RNA-seq 分析中使用单细胞或混合细胞需要从组织或培养物中分离细胞。尽管胰蛋白酶或最近的冷活性蛋白酶 (CAP) 已用于细胞解离,但基因表达变化被抑制的程度尚未阐明。为此,我们对小鼠骨骼肌祖细胞中酶依赖性基因表达变化进行了详细分析,重点关注酶处理时间、量和温度。我们发现通过酶处理改变表达的基因可以按时间依赖性方式分类,并且存在表达改变与酶处理时间无关的基因,量和温度。这项研究将作为基因的参考数据,这些基因应该被排除或考虑使用酶分离方法进行 RNA-seq 分析。
更新日期:2021-07-09
中文翻译:

骨骼成肌细胞酶解后基因表达变化的转录组分析
单细胞 RNA 测序分析是了解特定细胞状态的最有效工具之一。在 RNA-seq 分析中使用单细胞或混合细胞需要从组织或培养物中分离细胞。尽管胰蛋白酶或最近的冷活性蛋白酶 (CAP) 已用于细胞解离,但基因表达变化被抑制的程度尚未阐明。为此,我们对小鼠骨骼肌祖细胞中酶依赖性基因表达变化进行了详细分析,重点关注酶处理时间、量和温度。我们发现通过酶处理改变表达的基因可以按时间依赖性方式分类,并且存在表达改变与酶处理时间无关的基因,量和温度。这项研究将作为基因的参考数据,这些基因应该被排除或考虑使用酶分离方法进行 RNA-seq 分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号