Protein & Peptide Letters ( IF 1.0 ) Pub Date : 2021-07-31 , DOI: 10.2174/0929866528666210218215624 Salvador Eugenio C. Caoili 1
|
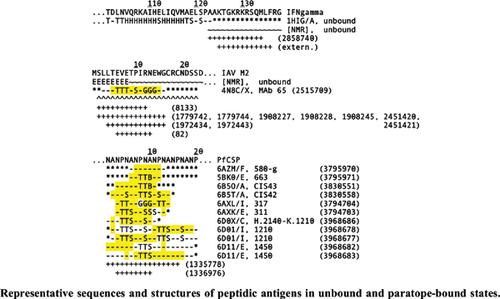
Background: B-cell epitope prediction is a computational approach originally developed to support the design of peptide-based vaccines for inducing protective antibody-mediated immunity, as exemplified by neutralization of biological activity (e.g., pathogen infectivity). Said approach is benchmarked against experimentally obtained data on paratope-epitope binding; but such data are curated primarily on the basis of immune-complex structure, obscuring the role of antigen conformational disorder in the underlying immune recognition process.
Objective: This work aimed to critically analyze the curation of epitope-paratope binding data that are relevant to B-cell epitope prediction for peptide-based vaccine design.
Methods: Database records on neutralizing monoclonal antipeptide antibody immune-complex structure were retrieved from the Immune Epitope Database (IEDB) and analyzed in relation to other data from both IEDB and external sources including the Protein Data Bank (PDB) and published literature, with special attention to data on conformational disorder among paratope-bound and unbound peptidic antigens.
Results: Data analysis revealed key examples of antipeptide antibodies that recognize conformationally disordered B-cell epitopes and thereby neutralize the biological activity of cognate targets (e.g., proteins and pathogens), with inconsistency noted in the mapping of some epitopes due to reliance on immune-complex structural details, which vary even among experiments utilizing the same paratope-epitope combination (e.g., with the epitope forming part of a peptide or a protein).
Conclusion: The results suggest an alternative approach to curating paratope-epitope binding data based on neutralization of biological activity by polyclonal antipeptide antibodies, with reference to immunogenic peptide sequences and their conformational disorder in unbound antigen structures.
中文翻译:

超越 B 细胞表位:整理关于抗肽互补位结合的阳性数据以支持基于肽的疫苗设计
背景:B 细胞表位预测是一种计算方法,最初开发用于支持基于肽的疫苗的设计,以诱导保护性抗体介导的免疫,例如生物活性(例如病原体感染性)的中和。所述方法以实验获得的关于互补位-表位结合的数据为基准;但这些数据主要是根据免疫复合物结构整理的,掩盖了抗原构象障碍在潜在免疫识别过程中的作用。
目的:这项工作旨在批判性地分析与基于肽的疫苗设计的 B 细胞表位预测相关的表位-互补位结合数据的管理。
方法:从免疫表位数据库(IEDB)中检索中和单克隆抗肽抗体免疫复合物结构的数据库记录,并结合来自 IEDB 和外部来源(包括蛋白质数据库(PDB)和已发表的文献)的其他数据进行分析,特别是注意互补位结合和未结合肽抗原中构象紊乱的数据。
结果:数据分析揭示了抗肽抗体的关键例子,它们识别构象紊乱的 B 细胞表位,从而中和同源靶标(例如,蛋白质和病原体)的生物活性,由于依赖免疫,在某些表位的映射中注意到不一致。复杂的结构细节,即使在使用相同互补位-表位组合的实验中也会有所不同(例如,表位形成肽或蛋白质的一部分)。
结论:结果提出了一种基于多克隆抗肽抗体对生物活性的中和,参考免疫原性肽序列及其在未结合抗原结构中的构象紊乱来管理互补位 - 表位结合数据的替代方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号