Computers & Graphics ( IF 2.5 ) Pub Date : 2021-05-07 , DOI: 10.1016/j.cag.2021.04.037 Khaled Al-Thelaya , Marco Agus , Nauman Ullah Gilal , Yin Yang , Giovanni Pintore , Enrico Gobbetti , Corrado Calí , Pierre J. Magistretti , William Mifsud , Jens Schneider
|
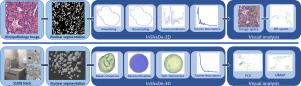
We present a shape processing framework for visual exploration of cellular nuclear envelopes extracted from microscopic images arising in histology and neuroscience. The framework is based on a novel shape descriptor of closed contours in 2D and 3D. In 2D, it relies on a geodesically uniform resampling of discrete curves to compute unsigned curvatures at vertices and edges based on discrete differential geometry. Our descriptor is, by design, invariant under translation, rotation, and parameterization. We achieve the latter invariance under parameterization shifts by using elliptic Fourier analysis on the resulting curvature vectors. Uniform scale-invariance is optional and is a result of scaling curvature features to z-scores. We further augment the proposed descriptor with feature coefficients obtained through sparse coding of the extracted cellular structures using K-sparse autoencoders. For the analysis of 3D shapes, we compute mean curvatures based on the Laplace-Beltrami operator on triangular meshes, followed by computing a spherical parameterization through mean curvature flow. Finally, we compute the Spherical Harmonics decomposition to obtain invariant energy coefficients. Our invariant descriptors provide an embedding into a fixed-dimensional feature space that can be used for various applications, e.g., as input features for deep and shallow learning techniques or as input for dimension reduction schemes to provide a visual reference for clustering shape collections. We demonstrate the capabilities of our framework in the context of visual analysis and unsupervised classification of 2D histology images and 3D nuclear envelopes extracted from serial section electron microscopy stacks.
中文翻译:

InShaDe:用于视觉 2D 和 3D 细胞和核形状分析和分类的不变形状描述符
我们提出了一个形状处理框架,用于对从组织学和神经科学中出现的显微图像中提取的细胞核包膜进行视觉探索。该框架基于 2D 和 3D 中闭合轮廓的新颖形状描述符。在 2D 中,它依赖于离散曲线的测地均匀重采样,以基于离散微分几何计算顶点和边缘处的无符号曲率。根据设计,我们的描述符在平移、旋转和参数化下是不变的。我们通过对结果曲率向量使用椭圆傅立叶分析,在参数化变化下实现了后者的不变性。统一尺度不变性是可选的,是将曲率特征缩放到 z 分数的结果。我们通过使用 K 稀疏自动编码器对提取的细胞结构进行稀疏编码获得的特征系数进一步增强了所提出的描述符。对于 3D 形状的分析,我们基于三角形网格上的 Laplace-Beltrami 算子计算平均曲率,然后通过平均曲率流计算球面参数化。最后,我们计算球谐分解以获得不变的能量系数。我们的不变描述符提供了一个嵌入到固定维度特征空间中的嵌入,该空间可用于各种应用,例如,作为深度和浅层学习技术的输入特征或作为降维方案的输入,为聚类形状集合提供视觉参考。











































 京公网安备 11010802027423号
京公网安备 11010802027423号