Current Pharmaceutical Biotechnology ( IF 2.8 ) Pub Date : 2021-08-31 , DOI: 10.2174/1389201021666201027154833 Clement Agoni 1 , Mahmoud E S Soliman 1
|
Aim: We seek to provide an understanding of the binding mechanism of Remdesivir, as well as structural and conformational implications on SARS-CoV-2 virus RNA-dependent RNA polymerase upon its binding and identify its crucial pharmacophoric moieties.
Background: The coronavirus disease of 2019 (COVID-19) pandemic had infected over a million people, with 65,000 deaths as of the first quarter of 2020. The current limitation of effective treatment options with no approved vaccine or targeted therapeutics for the treatment of COVID-19 has posed serious global health threats. This has necessitated several drug and vaccine development efforts across the globe. To date, the farthest in the drug development pipeline is Remdesivir.
Objectives: We performed the molecular dynamics simulation, quantified the energy contributions of binding site residues using per-residue energy decomposition calculations, and subsequently generated a pharmacophore model for the identification of potential SARS-CoV-2 virus RNA-dependent RNA polymerase inhibitors.
Methods: Integrative molecular dynamics simulations and thermodynamic calculations coupled with advanced post-molecular dynamics analysis techniques were employed.
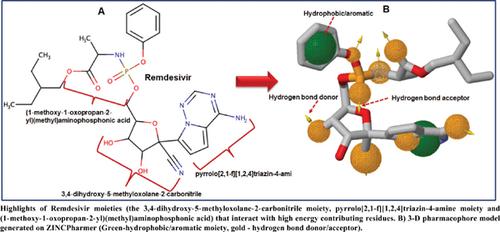
Results: Our analysis showed that the modulatory activity of Remdesivir is characterized by an extensive array of high-affinity and consistent molecular interactions with specific active site residues that anchor Remdemsivir within the binding pocket for efficient binding. These residues are ASP452, THR456, ARG555, THR556, VAL557, ARG624, THR680, SER681, and SER682. Results also showed that Remdesivir binding induces minimal individual amino acid perturbations, subtly interferes with deviations of C-α atoms, and restricts the systematic transition of SARS-CoV-2 RNA-dependent RNA polymerase from the “buried” hydrophobic region to the “surface-exposed” hydrophilic region. We also mapped a pharmacophore model based on the observed high-affinity interactions with SARSCoV- 2 virus RNA-dependent RNA polymerase, which showcased the crucial functional moieties of Remdesivir and was subsequently employed for virtual screening.
Conclusion: The structural insights and the provided optimized pharmacophoric model would augment the design of improved analogs of Remdesivir that could expand treatment options for COVID-19.
中文翻译:

瑞德西韦与 SARS-CoV-2 RNA 依赖性 RNA 聚合酶的结合可能为设计用于 COVID-19 治疗的潜在药物铺平道路
目的:我们试图了解 Remdesivir 的结合机制,以及对 SARS-CoV-2 病毒 RNA 依赖性 RNA 聚合酶结合后的结构和构象影响,并确定其关键的药效基团。
背景:截至 2020 年第一季度,2019 年冠状病毒病 (COVID-19) 大流行已感染超过 100 万人,死亡 65,000 人。目前有效治疗方案的局限性,尚无批准的疫苗或靶向疗法用于治疗 COVID -19 已构成严重的全球健康威胁。这需要在全球范围内进行多项药物和疫苗开发工作。迄今为止,药物开发管道中距离最远的是瑞德西韦。
目标:我们进行了分子动力学模拟,使用每个残基能量分解计算量化了结合位点残基的能量贡献,随后生成了一个药效团模型,用于识别潜在的 SARS-CoV-2 病毒 RNA 依赖性 RNA 聚合酶抑制剂。
方法:采用综合分子动力学模拟和热力学计算,并结合先进的后分子动力学分析技术。
结果:我们的分析表明,Remdesivir 的调节活性的特征在于与特定活性位点残基的广泛的高亲和力和一致的分子相互作用,这些残基将 Remdemsivir 锚定在结合口袋内以实现有效结合。这些残基是 ASP452、THR456、ARG555、THR556、VAL557、ARG624、THR680、SER681 和 SER682。结果还表明,Remdesivir 结合诱导最小的单个氨基酸扰动,巧妙地干扰 C-α 原子的偏差,并限制了 SARS-CoV-2 RNA 依赖性 RNA 聚合酶从“埋藏”疏水区域到“表面”的系统转变。 -暴露的”亲水区域。我们还根据观察到的与 SARS CoV-2 病毒 RNA 依赖性 RNA 聚合酶的高亲和力相互作用绘制了药效团模型,
结论:结构见解和提供的优化药效团模型将增强 Remdesivir 改进类似物的设计,从而扩大 COVID-19 的治疗选择。



























 京公网安备 11010802027423号
京公网安备 11010802027423号