Current Bioinformatics ( IF 2.4 ) Pub Date : 2021-01-31 , DOI: 10.2174/1574893615999200724145835 Dicle Yalcin 1 , Hasan H. Otu 1
|
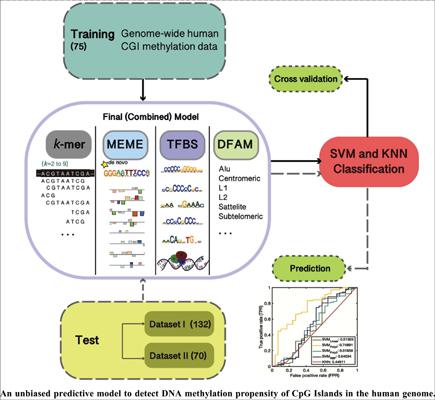
Background: Epigenetic repression mechanisms play an important role in gene regulation, specifically in cancer development. In many cases, a CpG island’s (CGI) susceptibility or resistance to methylation is shown to be contributed by local DNA sequence features.
Objective: To develop unbiased machine learning models–individually and combined for different biological features–that predict the methylation propensity of a CGI.
Methods: We developed our model consisting of CGI sequence features on a dataset of 75 sequences (28 prone, 47 resistant) representing a genome-wide methylation structure. We tested our model on two independent datasets that are chromosome (132 sequences) and disease (70 sequences) specific.
Results: We provided improvements in prediction accuracy over previous models. Our results indicate that combined features better predict the methylation propensity of a CGI (area under the curve (AUC) ~0.81). Our global methylation classifier performs well on independent datasets reaching an AUC of ~0.82 for the complete model and an AUC of ~0.88 for the model using select sequences that better represent their classes in the training set. We report certain de novo motifs and transcription factor binding site (TFBS) motifs that are consistently better in separating prone and resistant CGIs.
Conclusion: Predictive models for the methylation propensity of CGIs lead to a better understanding of disease mechanisms and can be used to classify genes based on their tendency to contain methylation prone CGIs, which may lead to preventative treatment strategies. MATLAB® and Python™ scripts used for model building, prediction, and downstream analyses are available at https://github.com/dicleyalcin/methylProp_predictor.
中文翻译:

检测人类基因组中CpG岛DNA甲基化倾向的无偏预测模型
背景:表观遗传抑制机制在基因调节中起着重要作用,特别是在癌症发展中。在许多情况下,CpG岛(CGI)的易感性或对甲基化的抗性被证明是由局部DNA序列特征引起的。
目的:分别开发针对不同生物学特征的无偏机器学习模型,这些模型可以预测CGI的甲基化倾向。
方法:我们在代表全基因组甲基化结构的75个序列(28个倾向,47个耐药)的数据集上开发了由CGI序列特征组成的模型。我们在两个独立的染色体(132个序列)和疾病(70个序列)特异性数据集中测试了我们的模型。
结果:与以前的模型相比,我们提供了更高的预测准确性。我们的结果表明,组合特征可以更好地预测CGI的甲基化倾向(曲线下面积(AUC)〜0.81)。我们的整体甲基化分类器在独立的数据集上表现良好,对于完整模型而言,其AUC约为〜0.82,对于模型而言,其AUC约为〜0.88,使用的选择序列可以更好地代表其在训练集中的类别。我们报告了某些从头开始的基序和转录因子结合位点(TFBS)的基序,它们在分离俯卧和耐药性CGI方面一直比较好。
结论:CGI甲基化倾向的预测模型可以使人们更好地了解疾病机理,并且可以根据基因倾向于含有甲基化倾向的CGI的趋势来对基因进行分类,这可能会导致预防性治疗策略。https://github.com/dicleyalcin/methylProp_predictor上提供了用于模型构建,预测和下游分析的MATLAB®和Python™脚本。











































 京公网安备 11010802027423号
京公网安备 11010802027423号