Current Bioinformatics ( IF 2.4 ) Pub Date : 2021-01-31 , DOI: 10.2174/1574893615999200623130416 Shicai Liu 1 , Hailin Tang 1 , Hongde Liu 1 , Jinke Wang 1
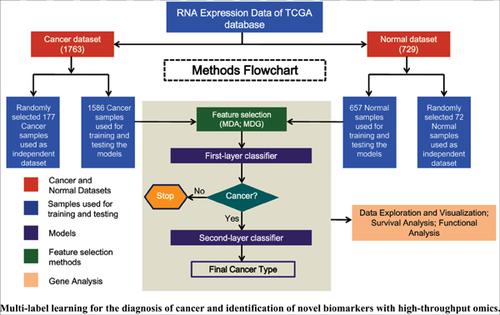
|
Background: The advancement of bioinformatics and machine learning has facilitated the diagnosis of cancer and the discovery of omics-based biomarkers.
Objective: Our study employed a novel data-driven approach to classifying the normal samples and different types of gastrointestinal cancer samples, to find potential biomarkers for effective diagnosis and prognosis assessment of gastrointestinal cancer patients.
Methods: Different feature selection methods were used, and the diagnostic performance of the proposed biosignatures was benchmarked using support vector machine (SVM) and random forest (RF) models.
Results: All models showed satisfactory performance in which Multilabel-RF appeared to be the best. The accuracy of the Multilabel-RF based model was 83.12%, with precision, recall, F1, and Hamming- Loss of 79.70%, 68.31%, 0.7357 and 0.1688, respectively. Moreover, proposed biomarker signatures were highly associated with multifaceted hallmarks in cancer. Functional enrichment analysis and impact of the biomarker candidates in the prognosis of the patients were also examined.
Conclusion: We successfully introduced a solid workflow based on multi-label learning with High- Throughput Omics for diagnosis of cancer and identification of novel biomarkers. Novel transcriptome biosignatures that may improve the diagnostic accuracy in gastrointestinal cancer are introduced for further validations in various clinical settings.
中文翻译:

多标签学习用于诊断癌症和鉴定具有高通量组学的新型生物标志物
背景:生物信息学和机器学习的发展促进了癌症的诊断和基于组学的生物标志物的发现。
目的:我们的研究采用一种新颖的数据驱动方法对正常样本和不同类型的胃肠道癌样本进行分类,以寻找可能有效地诊断和预测胃肠道癌患者的生物标志物。
方法:使用不同的特征选择方法,并使用支持向量机(SVM)和随机森林(RF)模型对所提出生物签名的诊断性能进行基准测试。
结果:所有模型均表现出令人满意的性能,其中Multilabel-RF似乎是最好的。基于Multilabel-RF的模型的准确性为83.12%,精度,召回率,F1和汉明损失分别为79.70%,68.31%,0.7357和0.1688。此外,提议的生物标志物签名与癌症中的多方面标志高度相关。还检查了功能富集分析和候选生物标志物对患者预后的影响。
结论:我们成功地引入了基于多标签学习和高通量组学的可靠工作流程,用于癌症诊断和新型生物标志物的鉴定。可以提高胃肠道癌症诊断准确性的新型转录组生物特征被引入,以在各种临床环境中进行进一步的验证。











































 京公网安备 11010802027423号
京公网安备 11010802027423号