Current Bioinformatics ( IF 2.4 ) Pub Date : 2021-01-31 , DOI: 10.2174/1574893615999200608130029 Akshatha Prasanna 1 , Vidya Niranjan 1
|
Background: Since bacteria are the earliest known organisms, there has been significant interest in their variety and biology, most certainly concerning human health. Recent advances in Metagenomics sequencing (mNGS), a culture-independent sequencing technology, have facilitated an accelerated development in clinical microbiology and our understanding of pathogens.
Objective: For the implementation of mNGS in routine clinical practice to become feasible, a practical and scalable strategy for the study of mNGS data is essential. This study presents a robust automated pipeline to analyze clinical metagenomic data for pathogen identification and classification.
Methods: The proposed Clin-mNGS pipeline is an integrated, open-source, scalable, reproducible, and user-friendly framework scripted using the Snakemake workflow management software. The implementation avoids the hassle of manual installation and configuration of the multiple commandline tools and dependencies. The approach directly screens pathogens from clinical raw reads and generates consolidated reports for each sample.
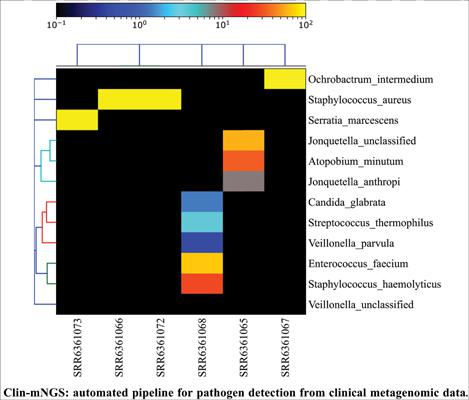
Results: The pipeline is demonstrated using publicly available data and is tested on a desktop Linux system and a High-performance cluster. The study compares variability in results from different tools and versions. The versions of the tools are made user modifiable. The pipeline results in quality check, filtered reads, host subtraction, assembled contigs, assembly metrics, relative abundances of bacterial species, antimicrobial resistance genes, plasmid finding, and virulence factors identification. The results obtained from the pipeline are evaluated based on sensitivity and positive predictive value.
Conclusion: Clin-mNGS is an automated Snakemake pipeline validated for the analysis of microbial clinical metagenomics reads to perform taxonomic classification and antimicrobial resistance prediction.
中文翻译:

Clin-mNGS:用于从临床基因组学数据检测病原体的自动化管道
背景:由于细菌是已知最早的生物,因此人们对其细菌的种类和生物学表现出极大的兴趣,其中最肯定的是与人类健康有关。Metagenomics测序(mNGS)的最新进展是一种独立于培养物的测序技术,它促进了临床微生物学的快速发展以及我们对病原体的理解。
目的:为了使mNGS在常规临床实践中的实施变得可行,对mNGS数据进行研究的实用且可扩展的策略至关重要。这项研究提出了一个强大的自动化管道来分析临床宏基因组学数据,以进行病原体鉴定和分类。
方法:拟议的Clin-mNGS管道是使用Snakemake工作流管理软件编写的,集成的,开源的,可伸缩的,可再现的和用户友好的框架。该实现避免了手动安装和配置多个命令行工具及依赖项的麻烦。该方法直接从临床原始读数中筛选病原体,并为每个样品生成合并报告。
结果:使用公开可用的数据演示了管道,并在桌面Linux系统和高性能集群上对其进行了测试。该研究比较了来自不同工具和版本的结果的变异性。这些工具的版本使用户可以修改。该管道可进行质量检查,过滤读取,宿主减法,组装重叠群,组装指标,细菌种类的相对丰度,抗菌素耐药基因,质粒发现和毒力因子鉴定。从管道中获得的结果将根据灵敏度和正预测值进行评估。
结论:Clin-mNGS是一条自动的Snakemake管道,经过验证可用于分析微生物临床宏基因组学读数,以进行分类学分类和抗菌素耐药性预测。











































 京公网安备 11010802027423号
京公网安备 11010802027423号