当前位置:
X-MOL 学术
›
Chem. Bio. Drug Des.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Computational design and modeling of nanobodies toward SARS-CoV-2 receptor binding domain
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2021-04-24 , DOI: 10.1111/cbdd.13847 Jingyi Yang 1 , Zhao Zhang 1 , Fengyuan Yang 1, 2 , Haiwei Zhang 3 , Haibo Wu 4 , Feng Zhu 1, 2 , Weiwei Xue 1
Chemical Biology & Drug Design ( IF 3.2 ) Pub Date : 2021-04-24 , DOI: 10.1111/cbdd.13847 Jingyi Yang 1 , Zhao Zhang 1 , Fengyuan Yang 1, 2 , Haiwei Zhang 3 , Haibo Wu 4 , Feng Zhu 1, 2 , Weiwei Xue 1
Affiliation
|
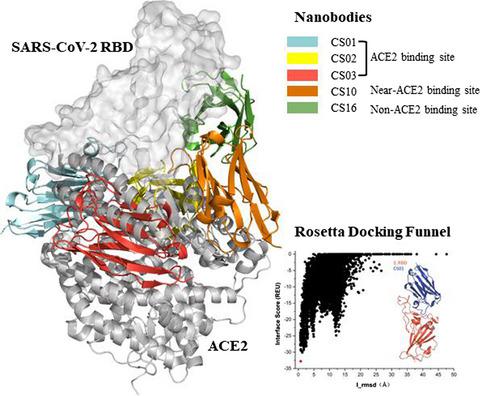
The ongoing pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global health concern and pose a serious threat to humanity. There is an urgent need for developing therapeutic drugs and (or) biologics to prevent the spread of the virus. The life cycle of SARS-CoV-2 shows that the virus enters host cells by first binding to angiotensin-converting enzyme 2 (ACE2) through its spike protein receptor-binding domain (RBD). Therefore, blocking the binding between of ACE2 and SARS-CoV-2 RBD can inhibit the virus infection in the host cells. In this study, by grafting the complementarity-determining regions (CDRs) of developed SARS-CoV, MERS-CoVs specific neutralizing antibodies (nAbs) include monoclonal antibodies (mAbs) as well as SARS-CoV-2 mAbs onto a known stable nanobody (Nb) scaffold, and a total of 16 Nbs sequences were designed. Five Nbs, namely CS01, CS02, CS03, CS10, and CS16, were selected based on the free energy landscape of protein docking verified by the recently reported Nb-RBD cocrystal structures. CS01, CS02, and CS03 occupied the ACE2 binding site of RBD, while CS10 and CS16 were proposed to inhibit the interaction between RBD and ACE2 through an allosteric mechanism. Based on the structures of the five Nbs in complex with RBD, seven brand-new Nbs with enhanced binding affinities (CS02_RD01, CS03_RD01, CS03_RD02, CS03_RD03, CS03_RD04, CS16_RD01, and CS16_RD02) were generated by redesign of residues on the interface of the five Nbs contact with SARS-CoV-2 RBD. In addition, the identified “hot spots” on the interface of each complex provide useful information to understand the binding mechanism of designed Nbs to SARS-CoV-2 RBD. In sum, the predicted stabilities and high binding affinities of the 11 (re)designed Nbs indicating the potential of the developed computational framework in this work to design effective agents to block the infection of SARS-CoV-2.
中文翻译:

针对 SARS-CoV-2 受体结合域的纳米抗体的计算设计和建模
由严重急性呼吸系统综合症冠状病毒 2 (SARS-CoV-2) 引起的 2019 年冠状病毒病 (COVID-19) 持续大流行已成为全球健康问题,并对人类构成严重威胁。迫切需要开发治疗药物和(或)生物制剂以防止病毒传播。SARS-CoV-2 的生命周期表明,该病毒首先通过其刺突蛋白受体结合域 (RBD) 与血管紧张素转换酶 2 (ACE2) 结合,从而进入宿主细胞。因此,阻断ACE2与SARS-CoV-2 RBD之间的结合可以抑制宿主细胞中的病毒感染。在这项研究中,通过嫁接发达的 SARS-CoV 的互补决定区 (CDR),MERS-CoVs 特异性中和抗体 (nAbs) 包括单克隆抗体 (mAbs) 以及 SARS-CoV-2 mAbs 到已知的稳定纳米抗体 (Nb) 支架上,总共设计了 16 个 Nbs 序列。根据最近报道的 Nb-RBD 共晶结构验证的蛋白质对接的自由能景观,选择了五个 Nb,即 CS01、CS02、CS03、CS10 和 CS16。CS01、CS02 和 CS03 占据 RBD 的 ACE2 结合位点,而 CS10 和 CS16 被提议通过变构机制抑制 RBD 和 ACE2 之间的相互作用。基于与 RBD 复合的五个 Nb 的结构,通过重新设计界面上的五个残基,生成了七个具有增强结合亲和力的全新 Nb(CS02_RD01、CS03_RD01、CS03_RD02、CS03_RD03、CS03_RD04、CS16_RD01 和 CS16_RD02) Nbs 与 SARS-CoV-2 RBD 的接触。此外,每个复合物界面上确定的“热点”为理解设计的 Nbs 与 SARS-CoV-2 RBD 的结合机制提供了有用的信息。总之,11 个(重新)设计的 Nbs 的预测稳定性和高结合亲和力表明,在这项工作中开发的计算框架有潜力设计有效的药物来阻止 SARS-CoV-2 的感染。
更新日期:2021-06-21
中文翻译:

针对 SARS-CoV-2 受体结合域的纳米抗体的计算设计和建模
由严重急性呼吸系统综合症冠状病毒 2 (SARS-CoV-2) 引起的 2019 年冠状病毒病 (COVID-19) 持续大流行已成为全球健康问题,并对人类构成严重威胁。迫切需要开发治疗药物和(或)生物制剂以防止病毒传播。SARS-CoV-2 的生命周期表明,该病毒首先通过其刺突蛋白受体结合域 (RBD) 与血管紧张素转换酶 2 (ACE2) 结合,从而进入宿主细胞。因此,阻断ACE2与SARS-CoV-2 RBD之间的结合可以抑制宿主细胞中的病毒感染。在这项研究中,通过嫁接发达的 SARS-CoV 的互补决定区 (CDR),MERS-CoVs 特异性中和抗体 (nAbs) 包括单克隆抗体 (mAbs) 以及 SARS-CoV-2 mAbs 到已知的稳定纳米抗体 (Nb) 支架上,总共设计了 16 个 Nbs 序列。根据最近报道的 Nb-RBD 共晶结构验证的蛋白质对接的自由能景观,选择了五个 Nb,即 CS01、CS02、CS03、CS10 和 CS16。CS01、CS02 和 CS03 占据 RBD 的 ACE2 结合位点,而 CS10 和 CS16 被提议通过变构机制抑制 RBD 和 ACE2 之间的相互作用。基于与 RBD 复合的五个 Nb 的结构,通过重新设计界面上的五个残基,生成了七个具有增强结合亲和力的全新 Nb(CS02_RD01、CS03_RD01、CS03_RD02、CS03_RD03、CS03_RD04、CS16_RD01 和 CS16_RD02) Nbs 与 SARS-CoV-2 RBD 的接触。此外,每个复合物界面上确定的“热点”为理解设计的 Nbs 与 SARS-CoV-2 RBD 的结合机制提供了有用的信息。总之,11 个(重新)设计的 Nbs 的预测稳定性和高结合亲和力表明,在这项工作中开发的计算框架有潜力设计有效的药物来阻止 SARS-CoV-2 的感染。











































 京公网安备 11010802027423号
京公网安备 11010802027423号