Environment International ( IF 10.3 ) Pub Date : 2021-04-13 , DOI: 10.1016/j.envint.2021.106556 Rongbin Xu 1 , Shuai Li 2 , Shanshan Li 1 , Ee Ming Wong 3 , Melissa C Southey 4 , John L Hopper 5 , Michael J Abramson 1 , Yuming Guo 1
|
Background
DNA methylation is a potential biological mechanism through which residential greenness affects health, but little is known about its association with greenness and whether the association could be modified by genetic background. We aimed to evaluate the association between surrounding greenness and genome-wide DNA methylation and potential gene-greenness interaction effects on DNA methylation.
Methods
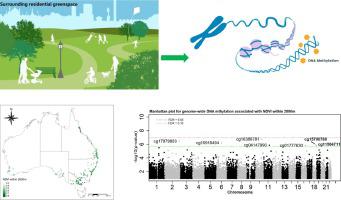
We measured blood-derived DNA methylation using the HumanMethylation450 BeadChip array (Illumina) for 479 Australian women, including 66 monozygotic, 66 dizygotic twin pairs, and 215 sisters of these twins. Surrounding greenness was represented by Normalized Difference Vegetation Index (NDVI) and Enhanced Vegetation Index (EVI) within 300, 500, 1000 or 2000 m surrounding participants’ home addresses. For each cytosine-guanine dinucleotide (CpG), the associations between its methylation level and NDVI or EVI were evaluated by generalized estimating equations, after adjusting for age, education, marital status, area-level socioeconomic status, smoking behavior, cell-type proportions, and familial clustering. We used comb-p and DMRcate to identify significant differentially methylated regions (DMRs). For each significant CpG, we evaluated the interaction effects of greenness and single-nucleotide polymorphisms (SNPs) within ±1 Mb window on its methylation level.
Results
We found associations between surrounding greenness and blood DNA methylation for one CpG (cg04720477, mapped to the promoter region of CNP gene) with false discovery rate [FDR] < 0.05, and for another 9 CpGs with 0.05 ≤ FDR < 0.10. For two of these CpGs, we found 33 SNPs significantly (FDR < 0.05) modified the greenness-methylation association. There were 35 significant DMRs related to surrounding greenness that were identified by both comb-p (Sidak p-value < 0.01) and DMRcate (FDR < 0.01). Those CpGs and DMRs were mapped to genes related to many human diseases, such as mental health disorders and neoplasms as well as nutritional and metabolic diseases.
Conclusions
Surrounding greenness was associated with blood DNA methylation of many loci across human genome, and this association could be modified by genetic variations.
中文翻译:

居住区周围的绿色和DNA甲基化:一个表观基因组范围的关联研究
背景
DNA甲基化是一种潜在的生物学机制,通过这种机制,住宅的绿色会影响健康,但对其与绿色的关联以及该关联是否可以通过遗传背景进行修饰的了解却很少。我们旨在评估周围的绿色和全基因组DNA甲基化之间的关联以及潜在的基因-绿色相互作用对DNA甲基化的影响。
方法
我们使用HumanMethylation450 BeadChip阵列(Illumina)对479名澳大利亚女性进行了血源性DNA甲基化测量,其中包括66对单卵双胞胎,66对卵双卵双胞胎以及这些双胞胎的215个姐妹。参与者的住所周围300、500、1000或2000 m内的归一化植被指数(NDVI)和增强植被指数(EVI)代表了周围的绿色。对于每个胞嘧啶-鸟嘌呤二核苷酸(CpG),在调整了年龄,教育程度,婚姻状况,地区水平的社会经济状况,吸烟行为,细胞类型比例后,通过广义估计方程对甲基化水平与NDVI或EVI之间的关联进行了评估。和家族聚类。我们使用梳状p和DMRcate来识别显着的差异甲基化区域(DMR)。对于每个重要的CpG,
结果
我们发现虚假发现率[FDR] <0.05的一个CpG(cg04720477,定位到CNP基因的启动子区域)和另外9个CpG的周围绿色与血液DNA甲基化之间的关联,假发现率[FDR] <0.05,而0.05≤FDR <0.10的另外9个CpGs。对于其中的两个CpG,我们发现33个SNP显着(FDR <0.05)修饰了绿色-甲基化缔合。通过梳状p(Sidak p值<0.01)和DMRcate(FDR <0.01)识别了35个与周围绿色相关的重要DMR。这些CpG和DMR被定位到与许多人类疾病相关的基因,例如精神健康疾病和肿瘤以及营养和代谢性疾病。
结论
周围的绿色与人类基因组中许多基因座的血液DNA甲基化有关,并且这种关联可以通过遗传变异进行修改。











































 京公网安备 11010802027423号
京公网安备 11010802027423号