Chinese Journal of Analytical Chemistry ( IF 1.2 ) Pub Date : 2021-04-08 , DOI: 10.1016/s1872-2040(21)60095-6 Jian-Bo TONG , Yi FENG , Tian-Hao WANG , Ding LUO
|
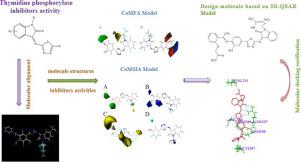
To obtain a better understanding of chemical biological interaction of isatin-based oxadiazole derivatives as a thymidine phosphorylase (TP) inhibitor, 30 kinds of quantitative structure activity relationship (QSAR) models of isatin-based oxadiazole derivatives were established using comparative molecular field analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA), respectively. Among them, the r2 and q2 of the best CoMFA model were 0.920 and 0.697, and the best CoMSIA model gave r2 = 0.912 and q2 = 0.692, respectively. Docking studies were used to find the actual conformations of chemicals in active sites of TP protease, as well as the binding pattern to the binding site in protease enzyme. The information provided by 3D-QSAR model and molecular docking offered useful references for the structural requirements of the 30 kinds of isatin-based oxadiazole derivatives and helped to design potential anti-tumor drugs of vascular inhibitors.
中文翻译:

基于Isatin的恶二唑衍生物作为胸苷磷酸化酶抑制剂的定量结构活性关系的研究
为了更好地了解基于靛红的恶二唑衍生物作为胸苷磷酸化酶(TP)抑制剂的化学生物学相互作用,使用比较分子场分析(CoMFA)建立了基于靛红的恶二唑衍生物的30种定量结构活性关系(QSAR)模型。 )和比较分子相似性指数分析(CoMSIA)。其中,最佳CoMFA模型的r 2和q 2为0.920和0.697,最佳CoMSIA模型的r 2 = 0.912和q 2分别为0.692。对接研究用于发现TP蛋白酶活性位点中化学物质的实际构象,以及与蛋白酶中结合位点的结合方式。3D-QSAR模型和分子对接所提供的信息为30种基于伊斯汀的恶二唑衍生物的结构要求提供了有用的参考,并有助于设计潜在的抗血管抑制剂的抗肿瘤药物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号