Structure ( IF 4.4 ) Pub Date : 2021-04-06 , DOI: 10.1016/j.str.2021.03.012 Betty W Shen 1 , Joel D Quispe 2 , Yvette Luyten 3 , Benjamin E McGough 4 , Richard D Morgan 3 , Barry L Stoddard 1
|
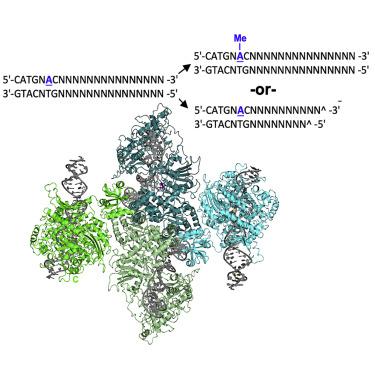
Restriction enzymes that combine methylation and cleavage into a single assemblage and modify one DNA strand are capable of efficient adaptation toward novel targets. However, they must reliably cleave invasive DNA and methylate newly replicated unmodified host sites. One possible solution is to enforce a competition between slow methylation at a single unmodified host target, versus faster cleavage that requires multiple unmodified target sites in foreign DNA to be brought together in a reaction synapse. To examine this model, we have determined the catalytic behavior of a bifunctional type IIL restriction-modification enzyme and determined its structure, via cryoelectron microscopy, at several different stages of assembly and coordination with bound DNA targets. The structures demonstrate a mechanism in which an initial dimer is formed between two DNA-bound enzyme molecules, positioning the endonuclease domain from each enzyme against the other's DNA and requiring further additional DNA-bound enzyme molecules to enable cleavage.
中文翻译:

通过 CryoEM 可视化的双功能限制性修饰酶协调噬菌体基因组降解与宿主基因组保护
将甲基化和切割结合成一个组合并修饰一条 DNA 链的限制酶能够有效适应新目标。然而,它们必须可靠地切割侵入性 DNA 并甲基化新复制的未修饰宿主位点。一种可能的解决方案是在单个未修饰的宿主靶标上进行缓慢的甲基化与需要在反应突触中将外来 DNA 中多个未修饰的靶位点聚集在一起的更快的切割之间的竞争。为了检查该模型,我们确定了双功能 IIL 型限制性修饰酶的催化行为,并通过低温电子显微镜确定了其在组装和与结合的 DNA 靶点协调的几个不同阶段的结构。











































 京公网安备 11010802027423号
京公网安备 11010802027423号