当前位置:
X-MOL 学术
›
Am. J. Primatol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploratory analysis reveals arthropod consumption in 10 lemur species using DNA metabarcoding
American Journal of Primatology ( IF 2.0 ) Pub Date : 2021-04-05 , DOI: 10.1002/ajp.23256 Amanda K Rowe 1 , Mariah E Donohue 2 , Elizabeth L Clare 3 , Rosie Drinkwater 3 , Andreas Koenig 1, 4 , Zachary M Ridgway 5 , Luke D Martin 6 , Eva S Nomenjanahary 7 , François Zakamanana 8 , Lovasoa J Randriamanandaza 8 , Thierry E Rakotonirina 8 , Patricia C Wright 1, 4
American Journal of Primatology ( IF 2.0 ) Pub Date : 2021-04-05 , DOI: 10.1002/ajp.23256 Amanda K Rowe 1 , Mariah E Donohue 2 , Elizabeth L Clare 3 , Rosie Drinkwater 3 , Andreas Koenig 1, 4 , Zachary M Ridgway 5 , Luke D Martin 6 , Eva S Nomenjanahary 7 , François Zakamanana 8 , Lovasoa J Randriamanandaza 8 , Thierry E Rakotonirina 8 , Patricia C Wright 1, 4
Affiliation
|
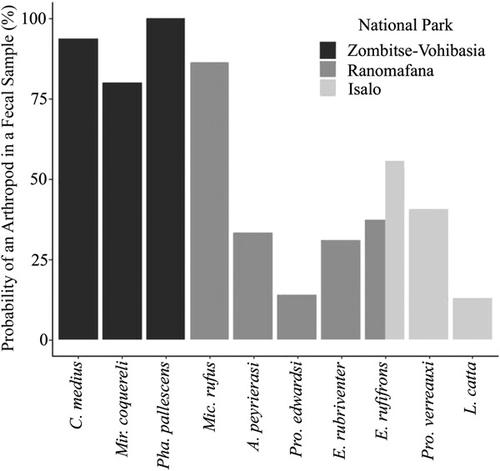
Arthropods (insects, spiders, etc.) can fulfill major nutritional requirements for primates, particularly in terms of proteins, fats, vitamins, and minerals. Yet, for many primate species we know very little about the frequency and importance of arthropod consumption. Traditional methods for arthropod prey identification, such as behavioral observations and fecal dissections, offer limited taxonomic resolution and, as a result, underestimate true diversity. Metabarcoding arthropod DNA from primate fecal samples provides a promising but underused alternative. Here, we inventoried arthropod prey diversity in wild lemurs by sequencing two regions of the CO1 gene. Samples were collected opportunistically from 10 species of lemurs inhabiting three national parks in southern Madagascar using a combination of focal animal follows and live trapping. In total, we detected arthropod DNA in 98 of the 170 fecal samples analyzed. Although all lemur species included in these analyses showed evidence of arthropod consumption, those within the family Cheirogaleidae appeared to consume the highest frequency and diversity of arthropods. To our knowledge, this study presents the first evidence of arthropod consumption in Phaner pallescens, Avahi peyrierasi, and Propithecus verreauxi, and identifies 32 families of arthropods as probable food items that have not been published as lemur dietary items to date. Our study emphasizes the importance of arthropods as a nutritional source and the role DNA metabarcoding can play in elucidating an animal's diet.
中文翻译:

探索性分析使用 DNA 元条形码揭示了 10 种狐猴物种的节肢动物消耗
节肢动物(昆虫、蜘蛛等)可以满足灵长类动物的主要营养需求,特别是在蛋白质、脂肪、维生素和矿物质方面。然而,对于许多灵长类动物,我们对节肢动物消费的频率和重要性知之甚少。传统的节肢动物猎物识别方法,如行为观察和粪便解剖,提供的分类分辨率有限,因此低估了真正的多样性。来自灵长类动物粪便样本的节肢动物 DNA Metabarcoding 提供了一种有前途但未充分利用的替代方法。在这里,我们通过对 CO1 基因的两个区域进行测序,盘点了野生狐猴的节肢动物猎物多样性。使用焦点动物追踪和现场诱捕的组合,从居住在马达加斯加南部三个国家公园的 10 种狐猴中随机收集样本。总的来说,我们在分析的 170 个粪便样本中的 98 个中检测到了节肢动物 DNA。尽管这些分析中包括的所有狐猴物种都显示出消耗节肢动物的证据,但 Cheirogaleidae 家族中的狐猴似乎消耗了最高频率和多样性的节肢动物。据我们所知,这项研究提供了节肢动物消费的第一个证据Phaner pallescens、Avahi peyrierasi和Propithecus verreauxi,并将 32 个节肢动物科确定为迄今为止尚未作为狐猴饮食项目发表的可能食物。我们的研究强调了节肢动物作为营养来源的重要性以及 DNA 元条形码在阐明动物饮食方面的作用。
更新日期:2021-05-28
中文翻译:

探索性分析使用 DNA 元条形码揭示了 10 种狐猴物种的节肢动物消耗
节肢动物(昆虫、蜘蛛等)可以满足灵长类动物的主要营养需求,特别是在蛋白质、脂肪、维生素和矿物质方面。然而,对于许多灵长类动物,我们对节肢动物消费的频率和重要性知之甚少。传统的节肢动物猎物识别方法,如行为观察和粪便解剖,提供的分类分辨率有限,因此低估了真正的多样性。来自灵长类动物粪便样本的节肢动物 DNA Metabarcoding 提供了一种有前途但未充分利用的替代方法。在这里,我们通过对 CO1 基因的两个区域进行测序,盘点了野生狐猴的节肢动物猎物多样性。使用焦点动物追踪和现场诱捕的组合,从居住在马达加斯加南部三个国家公园的 10 种狐猴中随机收集样本。总的来说,我们在分析的 170 个粪便样本中的 98 个中检测到了节肢动物 DNA。尽管这些分析中包括的所有狐猴物种都显示出消耗节肢动物的证据,但 Cheirogaleidae 家族中的狐猴似乎消耗了最高频率和多样性的节肢动物。据我们所知,这项研究提供了节肢动物消费的第一个证据Phaner pallescens、Avahi peyrierasi和Propithecus verreauxi,并将 32 个节肢动物科确定为迄今为止尚未作为狐猴饮食项目发表的可能食物。我们的研究强调了节肢动物作为营养来源的重要性以及 DNA 元条形码在阐明动物饮食方面的作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号