Computers & Electrical Engineering ( IF 4.0 ) Pub Date : 2021-03-30 , DOI: 10.1016/j.compeleceng.2021.107112 Amr Ezz El-Din Rashed , Marwa Obaya , Hossam El~Din Moustafa
|
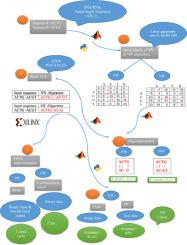
An optimized software and hardware digital implementation of two widely used DNA sequence alignment algorithms based on lookup table(LUT) is illustrated in this study. These algorithms are the best means for identifying similar regions between sequences. The proposed implementation relies on the complete parallelization of these foundational algorithms under certain limitations to overcome most of the problems of dynamic programming and hardware implementation. The proposed method takes O(N/4) calculation steps, where N is the length of each sequence with a minimum value of four (i.e., N = 4,8,12,…). A performance comparison between the state of art and our proposed algorithm is conducted for software and hardware implementation. Combinational circuits are used for FPGA-based hardware implementation of DNA sequence alignment algorithms. Performance and device resource usage are evaluated for different hardware designs. A customized convolution neural network model is used to implement global alignment and achieve 98.3% accuracy.
中文翻译:

使用FPGA和定制的卷积神经网络加速DNA配对序列比对
本文研究了基于查找表(LUT)的两种广泛使用的DNA序列比对算法的优化软件和硬件数字实现。这些算法是识别序列之间相似区域的最佳方法。所提出的实现依靠这些基础算法在某些限制下的完全并行化来克服动态编程和硬件实现中的大多数问题。所提出的方法需要O(N / 4)个计算步骤,其中N是每个序列的长度,最小值为4(即N = 4,8,12,...)。为了实现软件和硬件,对现有技术与我们提出的算法进行了性能比较。组合电路用于DNA序列比对算法的基于FPGA的硬件实现。针对不同的硬件设计评估性能和设备资源使用情况。使用定制的卷积神经网络模型来实现全局对齐并达到98.3%的准确性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号