Small Ruminant Research ( IF 1.8 ) Pub Date : 2021-03-31 , DOI: 10.1016/j.smallrumres.2021.106380 Reza Seyed Sharifi , Fatemeh.Ala Noshahr , Jamal Seifdavati , Nemat Hedayat Evrigh , Moises Cipriano-Salazar , Maria A. Mariezcurrena-Berasain
|
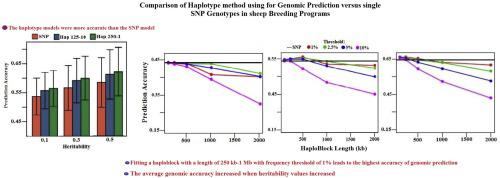
Linkage disequilibrium (LD) advancement map and the specification of population-level haplotype block structures are parameters that help manage the study of the Genome-wide Association (GWAS), and to comprehend the nature of the non-linear relationship among phenotypes and genotype. Compared with single nucleotide polymorphisms (SNP), genomic prediction fitting haplotype alleles and improve prediction accuracy; but the increase in accuracy belongs to how the Haplotype block is characterized. The Haplotype alleles were defined according to the SNP alleles in not covering blocks 125 Kb, 250 Kb, 500 Kb, and 1 Mb. The Haplotype alleles with frequencies below 1, 2.5, 5, or 10 % are eliminated. Using BayesB for genomic prediction fit covariate for SNP or haplotype. Three traits with three heritability levels of low: 0.10 (milk production (MILK)), medium: 0.30 (carcass weight (CARCASS)) and high: 0.45 (mature body weight (MATURE)) in sheep were assumed to be influencing the trait of interest. Under all situations, the average genomic accuracy increased when heritability values increased. When the haploblock length is properly characterized, covariates that are also suitable in haplotype alleles instead of SNPs will improve genome precision. Fitting a haploblock with a length of 250 kb-1 Mb with a frequency threshold of 1% leads to the highest accuracy of genomic prediction. Increasing the frequency threshold had a more negative impact on both accuracy and bias for the longer haploblocks than shorter haploblocks.
中文翻译:

绵羊育种计划中用于基因组预测的单倍型方法与单SNP基因型的比较
连锁不平衡(LD)进阶图和群体水平单倍型块结构的规范是有助于管理全基因组协会(GWAS)的研究的参数,并有助于理解表型和基因型之间非线性关系的性质。与单核苷酸多态性(SNP)相比,基因组预测适合单倍型等位基因并提高预测准确性;但是准确性的提高属于单倍型模块的特征。根据SNP等位基因在不覆盖125 Kb,250 Kb,500 Kb和1 Mb的区域中定义了单倍型等位基因。消除频率低于1、2.5、5或10%的单倍型等位基因。使用BayesB进行基因组预测适合SNP或单倍型的协变量。三个性状水平较低的三个特征:0.10(产奶量(MILK)),中等:绵羊中的0.30(car体重量(CARCASS))和高:0.45(成熟体重(MATURE))影响了所关注的性状。在所有情况下,当遗传度值增加时,平均基因组准确性就会增加。当适当地表征了单倍体长度时,也适用于单倍型等位基因而不是SNP的协变量将提高基因组精度。拟合长度为250 kb-1 Mb且频率阈值为1%的单倍型可导致基因组预测的最高精确度。频率阈值的增加对较长单倍单元的准确性和偏差产生的负面影响要比较短单倍单元的负面影响更大。当遗传力值增加时,平均基因组准确性增加。当适当地表征了单倍体长度时,也适用于单倍型等位基因而不是SNP的协变量将提高基因组精度。拟合长度为250 kb-1 Mb且频率阈值为1%的单倍型可导致基因组预测的最高准确度。频率阈值的增加对较长单倍单元的准确性和偏差产生的负面影响要比较短单倍单元的负面影响更大。当遗传力值增加时,平均基因组准确性增加。当适当地表征了单倍体长度时,也适用于单倍型等位基因而不是SNP的协变量将提高基因组精度。拟合长度为250 kb-1 Mb且频率阈值为1%的单倍型可导致基因组预测的最高准确度。频率阈值的增加对较长单倍单元的准确性和偏差产生的负面影响要比较短单倍单元的负面影响更大。



























 京公网安备 11010802027423号
京公网安备 11010802027423号