Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2021-03-21 , DOI: 10.1016/j.jmgm.2021.107902 G I Makarov 1 , R V Reshetnikova 1
|
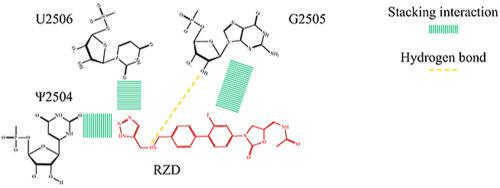
Radezolid is a promising antibiotic of oxazolidinone family, which is able to overcome effect of some linezolid resistance mechanisms of bacterial ribosomes. The structure of the radezolid complex with ribosomes was never published but, by analogy with linezolid, it is considered to prevent the binding of aminoacyl–tRNA to the A–site of the ribosome large subunit. However, as with linezolid, it can be assumed that radezolid binds to the alternative binding site existing in the A,A/P,P–ribosome. In the present article we have investigated this issue by molecular dynamics simulations and proposed the structure of the radezolid complex with a E. coli ribosome, which is consistent with available data of biochemical investigations of radezolid action.
中文翻译:

雷克唑利德与非规范氯霉素结合位点的相互作用的分子动力学模拟研究
Radezolid是恶唑烷酮家族的一种有前途的抗生素,它能够克服细菌核糖体对某些利奈唑胺耐药机制的作用。雷地唑烷具有核糖体的复合物的结构从未公开,但与利奈唑胺类似,它被认为可以防止氨酰基tRNA与核糖体大亚基的A位点结合。但是,与利奈唑胺一样,可以假定雷地唑利德与存在于A,A / P,P-核糖体的替代结合位点结合。在本文中,我们已通过分子动力学模拟研究了这一问题,并提出了雷迪唑利德与大肠杆菌核糖体复合物的结构,这与雷迪唑利德作用的生化研究的现有数据一致。











































 京公网安备 11010802027423号
京公网安备 11010802027423号