Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2021-03-23 , DOI: 10.1016/j.jmgm.2021.107906 Igor Lima 1 , Elio A Cino 1
|
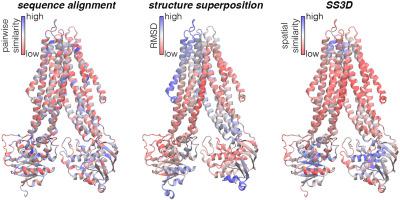
Homologous proteins are often compared by pairwise sequence alignment, and structure superposition if the atomic coordinates are available. Unification of sequence and structure data is an important task in structural biology. Here, we present the Sequence Similarity 3D (SS3D) method of integrating sequence and structure information. SS3D is a distance and substitution matrix-based method for straightforward visualization of regions of similarity and difference between homologous proteins. This work details the SS3D approach, and demonstrates its utility through case studies comparing members of several protein families. The examples show that SS3D can effectively highlight biologically important regions of similarity and dissimilarity. We anticipate that the method will be useful for numerous structural biology applications, including, but not limited to, studies of binding specificity, structure-function relationships, and evolutionary pathways. SS3D is available with a manual and tutorial at https://github.com/0x462e41/SS3D/.
中文翻译:

用于蛋白质家族比较的3D序列相似性
同源蛋白质通常通过成对序列比对和结构重叠(如果原子坐标可用)进行比较。序列和结构数据的统一是结构生物学中的重要任务。在这里,我们提出了将序列和结构信息整合在一起的序列相似性3D(SS3D)方法。SS3D是一种基于距离和置换矩阵的方法,用于直观显示同源蛋白之间相似性和差异性的区域。这项工作详细介绍了SS3D方法,并通过比较几个蛋白质家族成员的案例研究证明了其实用性。这些示例表明,SS3D可以有效地突出相似性和相异性的重要生物学区域。我们预计该方法将可用于众多结构生物学应用,包括:但不限于结合特异性,结构-功能关系和进化途径的研究。可在https://github.com/0x462e41/SS3D/获得手册和教程的SS3D。











































 京公网安备 11010802027423号
京公网安备 11010802027423号